Figure 3. TERT activates ERVs by interacting with Sp1.
-
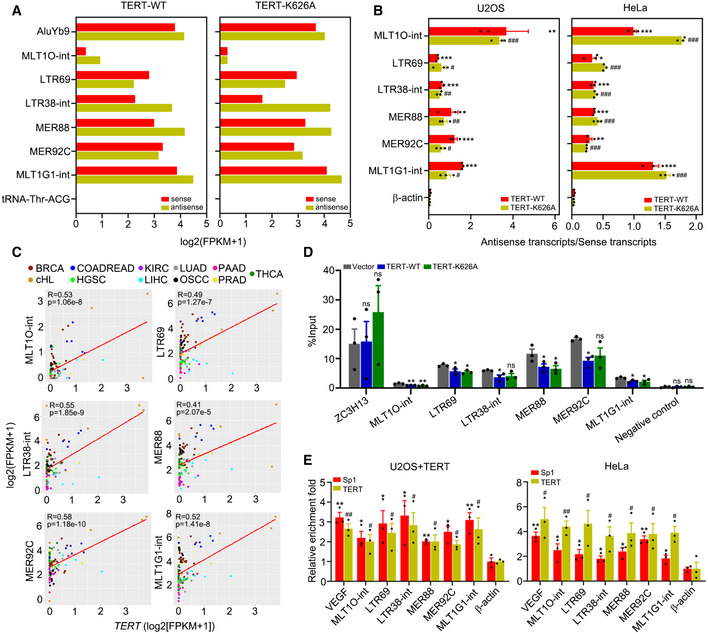
AIdentification of dsRNAs activated by TERT‐WT or TERT‐K626A using dsRIP‐seq in U2OS cells. AluYb9 and tRNA‐Thr‐ACG as positive and negative controls, respectively.
-
BQuantification of antisense/sense transcripts of ERVs activated by TERT‐WT or TERT‐K626A in U2OS and HeLa cells using strand‐specific RT–qPCR.
-
CRegression analysis of the correlation between TERT and TA‐ERVs expression in 103 tumour samples across 11 cancer types. BRCA (breast invasive carcinoma), n = 17. cHL (classical Hodgkin lymphoma), n = 5. COADREAD (colon adenocarcinoma/rectum adenocarcinoma), n = 6. HGSC (high‐grade serous ovarian cancer), n = 6. KIRC (kidney renal clear cell carcinoma), n = 12. LIHC (liver hepatocellular carcinoma), n = 9. LUAD (lung adenocarcinoma), n = 6. OSCC (oral squamous cell carcinoma), n = 8. PAAD (pancreatic adenocarcinoma), n = 10. PRAD (prostate adenocarcinoma), n = 6. THCA (thyroid carcinoma), n = 18.
-
DMeDIP‐qPCR analysis of TA‐ERVs sites in HeLa cells ectopically expressing TERT‐WT or TERT‐K626A. Primer sets recognizing exon 8 of the human ZC3H13 gene and a gene desert on chromosome 12 were used for positive and negative controls, respectively.
-
EChIP‐qPCR for TERT and Sp1 targets on TA‐ERVs in U2OS cells with TERT ectopic expression and in HeLa cells. β‐actin promoter was used as a negative control, and VEGF promoter was used as a positive control.
Data information: Data represent mean ± SEM of three biological replicates (B, D and E). */# P < 0.05, **/## P < 0.01, ***/### P < 0.001, ns, not significant. Unpaired two‐tailed Student’s t‐tests were used, and asterisks indicating significance for each ERV versus β‐actin in TERT‐WT while hashtags indicating significance for each ERV versus β‐actin in TERT‐K626A in (B) and (E), one‐way ANOVA with Fisher's LSD tests was used for (D).
