Figure 4. 4D7 has no effect on normal progenitor cells and shows activity against ruxolitinib‐resistant cells without haematological toxicity.
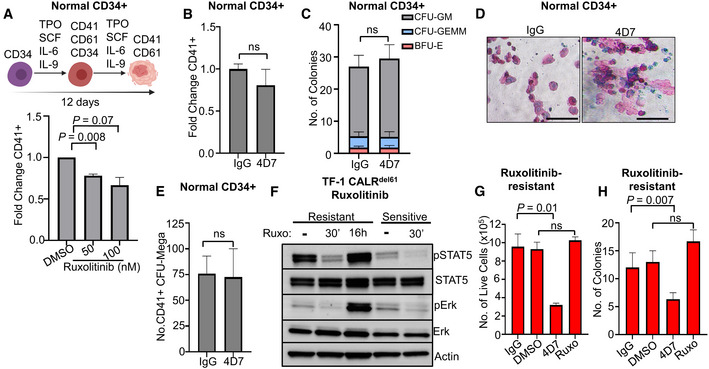
- Graph showing effect of ruxolitinib on megakaryocyte differentiation at 50 and 100 nM in healthy CD34+ cells (n = 4 different cords collected with three technical replicates).
- Effect of 4D7 on TPO‐dependent megakaryocyte differentiation of healthy CD34+ cells cultured with 20 µg/ml 4D7 or IgG control (n = 2 biological replicates, with two technical replicates).
- Total numbers of haematopoietic colonies plated in MethoCult from healthy cord blood after treatment with 4D7 or IgG. CFU‐GM, colony forming unit–granulocyte macrophage; BFU‐E, blast forming unit–erythroid; CFU‐GEMM, colony forming unit–granulocyte, erythroid, monocyte and megakaryocyte (n = 3 biological replicates).
- CD41+ megakaryocyte colonies from healthy CD34+ cells observed on collagen matrix after 4D7 treatment. Colonies were cultured in TPO, SCF, IL‐9 and IL‐3. Scale bar indicates 100 µm.
- Number of megakaryocyte colony forming units observed in MegaCult assay after treatment of healthy cord blood CD34+ cells with either IgG or 4D7 (n = 3 biological replicates).
- Western blot showing signalling in ruxolitinib‐resistant TF‐1 TpoR CALRdel61 compared to ruxolitinib‐sensitive TF‐1 TpoR CALRdel61 cells after treatment with 100 nM ruxolitinib for 16 h or 30 min and blotted for phospho‐STAT5, total STAT5, phospho‐ERK, total ERK and actin as indicated. Ruxolitinib‐sensitive cells were non‐viable after 16 h of treatment.
- Comparison of cell growth after DMSO, 20 µg/ml 4D7, IgG or 100 nM ruxolitinib treatment over 4 days of ruxolitinib‐resistant TF‐1 TpoR CALRdel61. Cells counted using trypan blue exclusion (n = 3 biological replicates with three technical replicates).
- Number of colonies observed from cells plated in MethoCult following 72 h of treatment with DMSO, 20 µg/ml 4D7, IgG or 100 nM ruxolitinib performed in ruxolitinib‐resistant TF‐1 TpoR CALRdel61 cells (n = 3 biological replicates with three technical replicates).
Data information: Error bars on (A, B, C and E) indicate standard error of mean. Error bars on (G and H) represent standard deviation. All P‐values are unpaired Student's unpaired t‐test.
Source data are available online for this figure.
