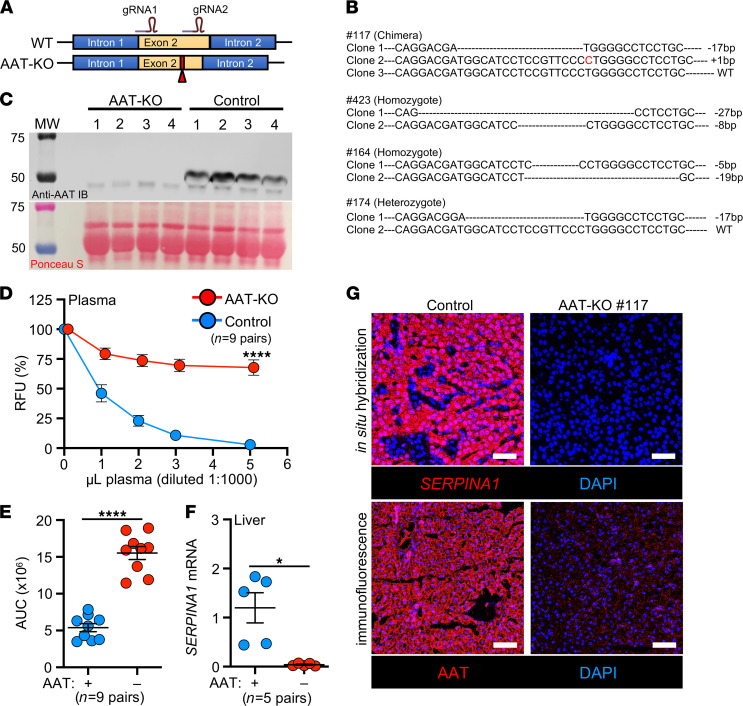
Figure 1. Generation and validation of an AAT-KO ferret.
(A) Schematic for Cas9-mediated disruption of the SERPINA1 locus in ferret zygotes. (B) Sequences of founders screened for expression, with the insertion and deletion polymorphisms they harbored depicted. (C) Western blot of plasma AAT protein in 4 AAT-KO ferrets and matched WT controls. Ponceau S–stained loading control is shown below. All draws were obtained at approximately 2 months old. (D) Inhibitory capacity of NE in increasing volumes of plasma from AAT-KO and control ferrets (n = 9 pairs; P value by mixed effects model, P = 1.6 × 10–5). (E) Quantification of NE inhibitory capacity based on AUC for each genotype in D (n = 9 pairs; P value by Student’s t test, P = 2.9 × 10–7). (F) Quantification of SERPINA1 mRNA expression in liver, normalized to GAPDH (n = 5 pairs; P value by Student’s t test, P < 0.05). (G) Localization of SERPINA1 transcripts and AAT protein in liver sections from a representative matched pair. Nuclei are counterstained with DAPI (blue). Scale bars: 50 μm. All graphs show mean ± SEM; some error bars are hidden by symbols. *P < 0.05; ****P < 0.0001.