Figure 2.
PredictProtein results
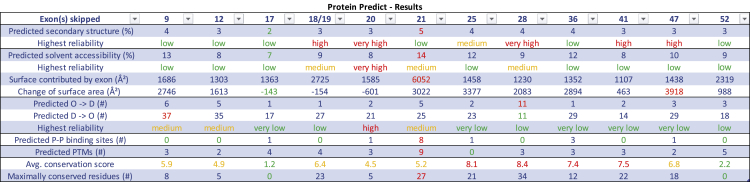
Summary of in silico analysis for selected exons. Exon(s) skipped: number of exons skipped according to continuous 1…58 exon numbering outlined in LRG_214. Predicted secondary structure (%): the percentage of residues in the remaining protein (NF1delEX) that are predicted to undergo a change in secondary structure when compared with (full-length) human neurofibromin. Highest reliability (secondary structure): predictions of secondary structures have a reliability score assigned. Here we report the highest reliability reported for any such prediction. Predicted solvent accessibility (%): the percentage of residues in the remaining protein (NF1delEX) that are predicted to undergo a change in solvent accessibility when compared with predictions for (full-length) human neurofibromin. Highest reliability (solvent accessibility): predictions of solvent accessibility have a reliability score assigned. Here we report the highest reliability reported for any prediction. Surface contributed by exon (Å2): predicted solvent accessibility in squared angstroms attributed to the amino acids that have been translated from the exon(s). Change of surface area (Å2): predicted total solvent accessibility in squared angstroms of human full-length (fl) neurofibromin minus the predicted surface area contributed by the skipped exon(s) minus the predicted solvent accessibility of the protein with skipped exon. A positive value means that the surface of fl is larger than the surface of the protein with the skipped exon. Predicted O → D (#): number of residues in the shortened protein (NF1delEX) predicted to change status from ordered to disordered, when compared with full-length neurofibromin. Predicted D → O (#): number of residues in the shortened protein (NF1delEX) predicted to change status from disordered to ordered, when compared with full-length neurofibromin. Highest reliability (ordered/disordered): predictions of status (ordered versus disordered) have a reliability score assigned. Here we report the highest reliability reported for any prediction. Predicted P-P binding sites (#): number of predicted protein-protein binding sites as predicted by PROFisis (part of PredictProtein). Predicted PTMs: number of PTMs as predicted from Prosite as part of PredictProtein, CKSAAP_UbSite, and UbiProber (with score >0.8). This also includes PTMs that are not on residues formed by the exon but likely affected by the exon skipping, if the recognition sequence is directly adjacent to the skipped region. Avg. conservation score: average conservation score for each exon as calculated by ConSurf. Maximally conserved residues (#): number of amino acids with the highest conservation score for each exon as obtained from ConSurf.