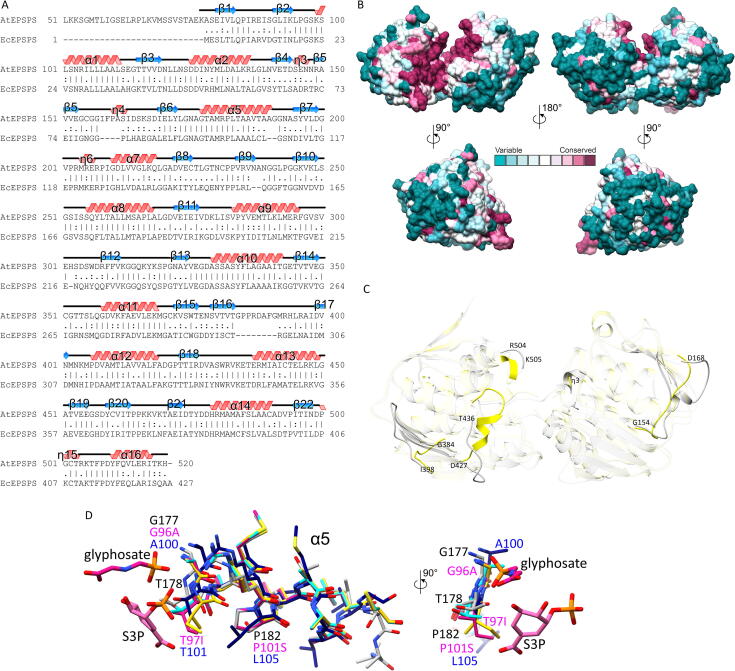
Fig. 5.
Comparison of AtEPSPS with its homologues from other species. Panel A presents the sequence alignment of AtEPSPS with EcEPSPS; secondary structure elements are shown for this work structure. Residue conservation in EPSPS homologs sharing between 35% and 95% sequence identity to AtEPSPS is shown in panel B, color-coded according to the key. Panel C presents the structural comparison of this work AtEPSPS (gray) structure with EcEPSPS (yellow, PDB ID 1g6s [45]). Due to the different conformation in the 1g6s structure, the terminal and central domains were superposed and compared independently. Fragments that overlap well are semitransparent to highlight the differences. Residue numbering is given for AtEPSPS. Superposition of the structures of glyphosate insensitive EPSPS variants onto the AtEPSPS structure is shown in panel D. The central domains were superposed but only the α5 helix (in two views) is visualized for clarity. Residue substitutions tested in EcEPSPS are labeled in magenta while corresponding positions in AtEPSPS and CP4 EPSPS are black and navy blue, respectively. The compared structures of EcEPSPS are: G96A mutant (cyan, PDB ID: 1mi4, [51]); T97I (pink, 3fjz, [50]); T97I/P101S (yellow, 3fko, [50]); CP4 EPSPS (navy blue, 2gg6, [52]). Glyphosate and S3P originate from the 3fjz structure. The view in panel D (left) is similar to that in C. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)