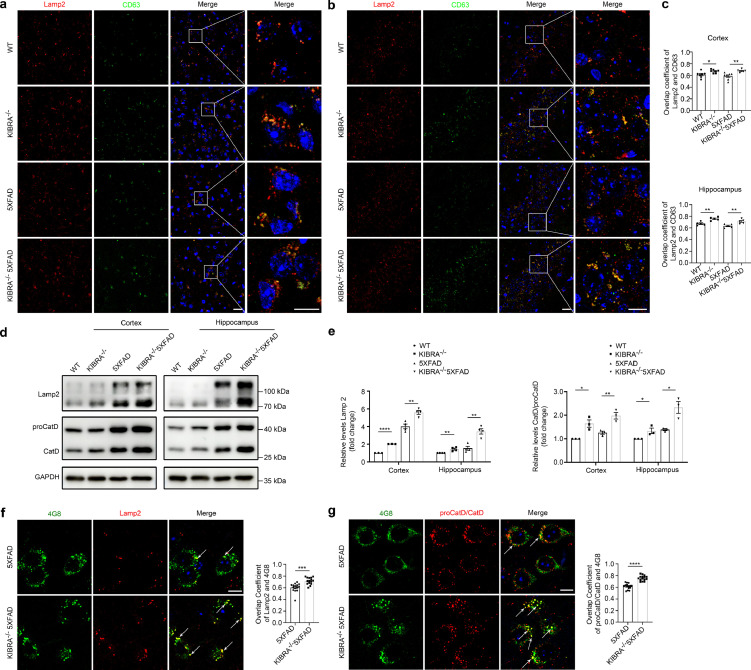
Figure 5.
KIBRA Knockout leads to AβPP/Aβ aggregation and degradation by lysosomes. (a and b) Confocal microscopy analysis of MVBs marker CD63 (green) and lysosomal marker LAMP2 (red) co-localization in cortical layers 5 (a) and hippocampal CA1-3 (b) regions of WT, KIBRA−/−, 5XFAD, and KIBRA−/−5XFAD mice. Nuclei were stained with DAPI. Scale bar in left panel = 20 µm. Scale bar in right panel = 10 µm. (c) Quantification analysis of overlap coefficient per view (n = 7 in the cortex and n = 5 in the hippocampus). (d) WB analysis of lysosomal marker Lamp2 and Cathepsin D levels in brain tissue lysates from cortical and hippocampal regions of 3–5-month-old mice. (e) Quantification analysis of lysosomal membrane protein levels (Lamp2, left panel) and ratio of CatD/proCatD (right panel) in cortical and hippocampal regions from 3–5-month-old mice of the indicated genotypes. Quantification results were plotted as dot plots, showing the mean ± SE of at least three independent experiments. *P < 0.05, ⁎⁎P < 0.01, ⁎⁎⁎⁎P < 0.0001 as determined by a two-tailed t test. (f) Confocal microscopy analysis of lysosomal membrane marker Lamp2 (red) with Aβ/AβPP (anti-Aβ, clone 4G8, green) in brain slice of 5XFAD and KIBRA−/−5XFAD mice. Scale bar = 10 µm. The overlap coefficient per cell was quantified (n = 15). (g) Confocal microscopy analysis of Cathepsin D (red) with Aβ/AβPP (anti-Aβ, clone 4G8, green) in brain slice of 5XFAD and KIBRA−/−5XFAD mice. Scale bar = 10 µm. Overlap coefficient per cell was quantified (n = 15). f-g. Quantification result were plotted as dot plots, showing the mean ± SE. ⁎⁎⁎P < 0.001, ⁎⁎⁎⁎P < 0.0001 as determined by a two-tailed student's t test.