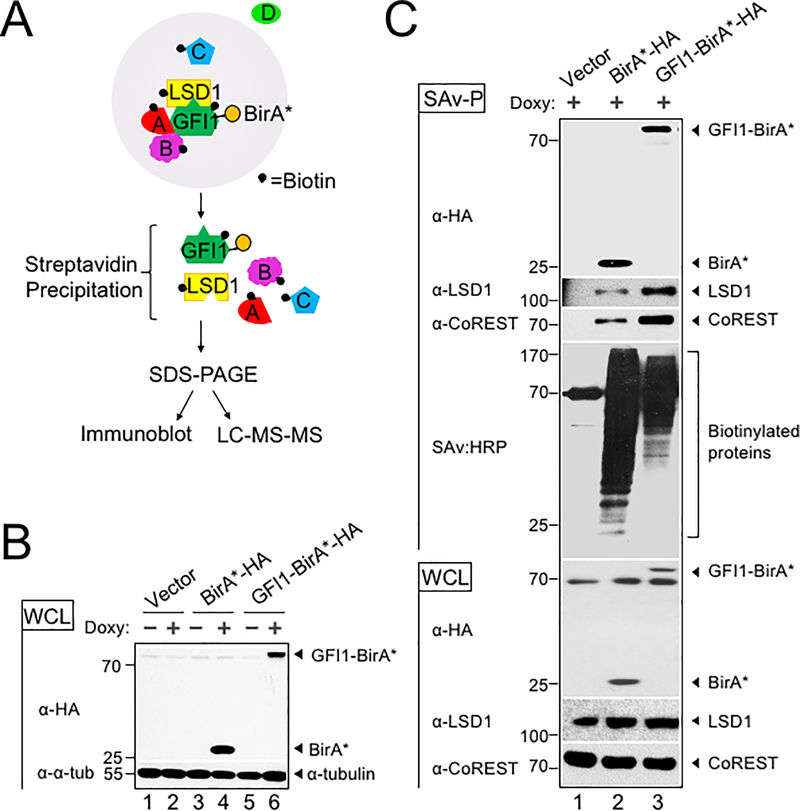
Figure 1.
BioID proximity labeling identifies GFI1-interacting proteins. (A) Scheme for BioID proximity labeling method to identify GFI1-proximate proteins. Gray circle reflects the radius for biotinylation of nearby molecules such as GFI1 itself and its known binding partner LSD1, as well as unknown proteins A, B and C but not D. (B) CCRF-CEM cells inducibly express BirA*-HA alone or as a fusion with GFI1. Biotin-treated CCRF-CEM cells transduced with empty vector (lanes 1–2), BirA*-HA (lanes 3–4), or GFI1-BirA*-HA (lanes 5–6) were treated with doxycycline (Doxy) (+) or vehicle control (−). Whole cell lysates (WCL) were resolved by SDS-PAGE and immunoblotted using HA antibodies. α-tubulin is shown as a loading control. (C) Biotinylation of GFI1-associated proteins LSD1 and CoREST is enriched using GFI1-BirA*-HA expressing CCRF-CEM cells relative to empty vector or BirA*-HA controls. Whole cell lysates prepared as in (B) were used for immunoblotting or were first precipitated using streptavidin Sepharose beads (SAv-P), and subsequently analyzed by immunoblotting with anti-HA, anti-LSD1 or anti-CoREST antibodies, or streptavidin-HRP (SAv:HRP) to detect biotinylation.