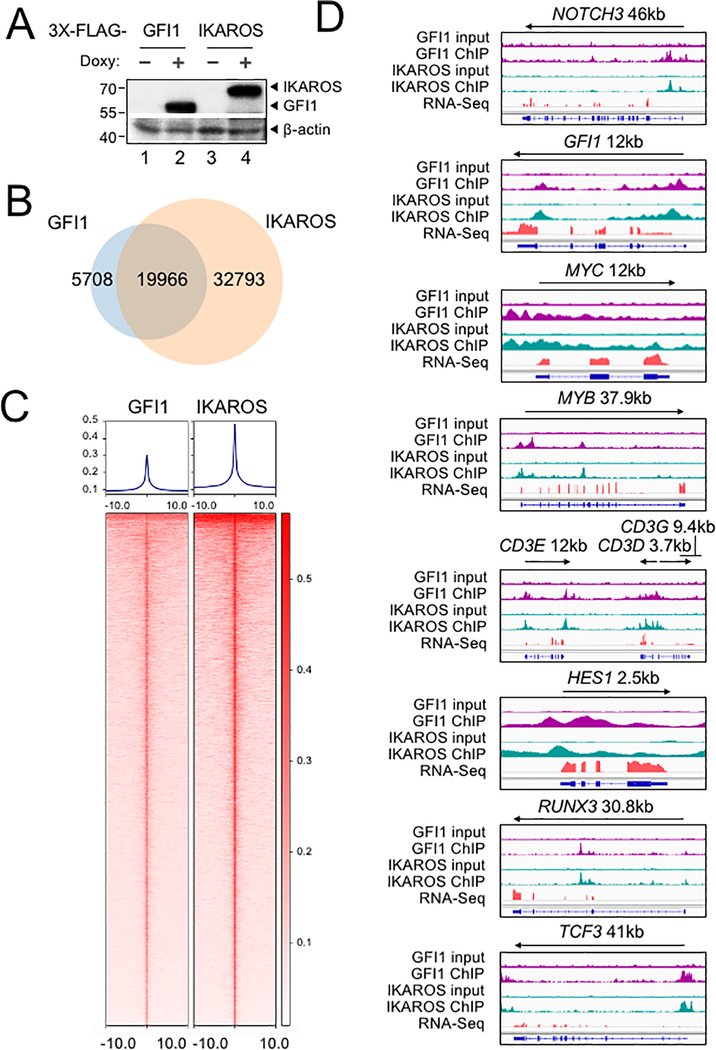
Figure 4.
Identification of GFI1 and IKAROS targets in CCRF-CEM cells. (A) Validation of GFI1 and IKAROS expression in CCRF-CEM cell lines by Western blotting. CCRF-CEM cells expressing GFI1–3×FLAG and IKAROS-3×FLAG were used for ChIP-Seq. (B) Venn diagram showing enumeration of unique and shared GFI1 and IKAROS ChIP-Seq peaks. (C) Left: GFI1 ChIP-Seq heatmap showing all peak sites, centered by the ChIP peak summit and arranged from strongest to weakest. Right: the same set of peak summits in the same order shown as an IKAROS binding heatmap. (D) Example Integrative Genomics Viewer (IGV) tracks of GFI1 ChIP-Seq and input controls (purple), IKAROS ChIP-Seq and input controls (teal), together with previously published RNA-Seq (peach) of CCRF-CEM cells. NOTCH3, GFI1, MYC, MYB, CD3, HES1, RUNX3 and TCF3 tracks are shown.