Figure 1. SQLE is overexpressed and correlated with poor survival in BC and NSCLC.
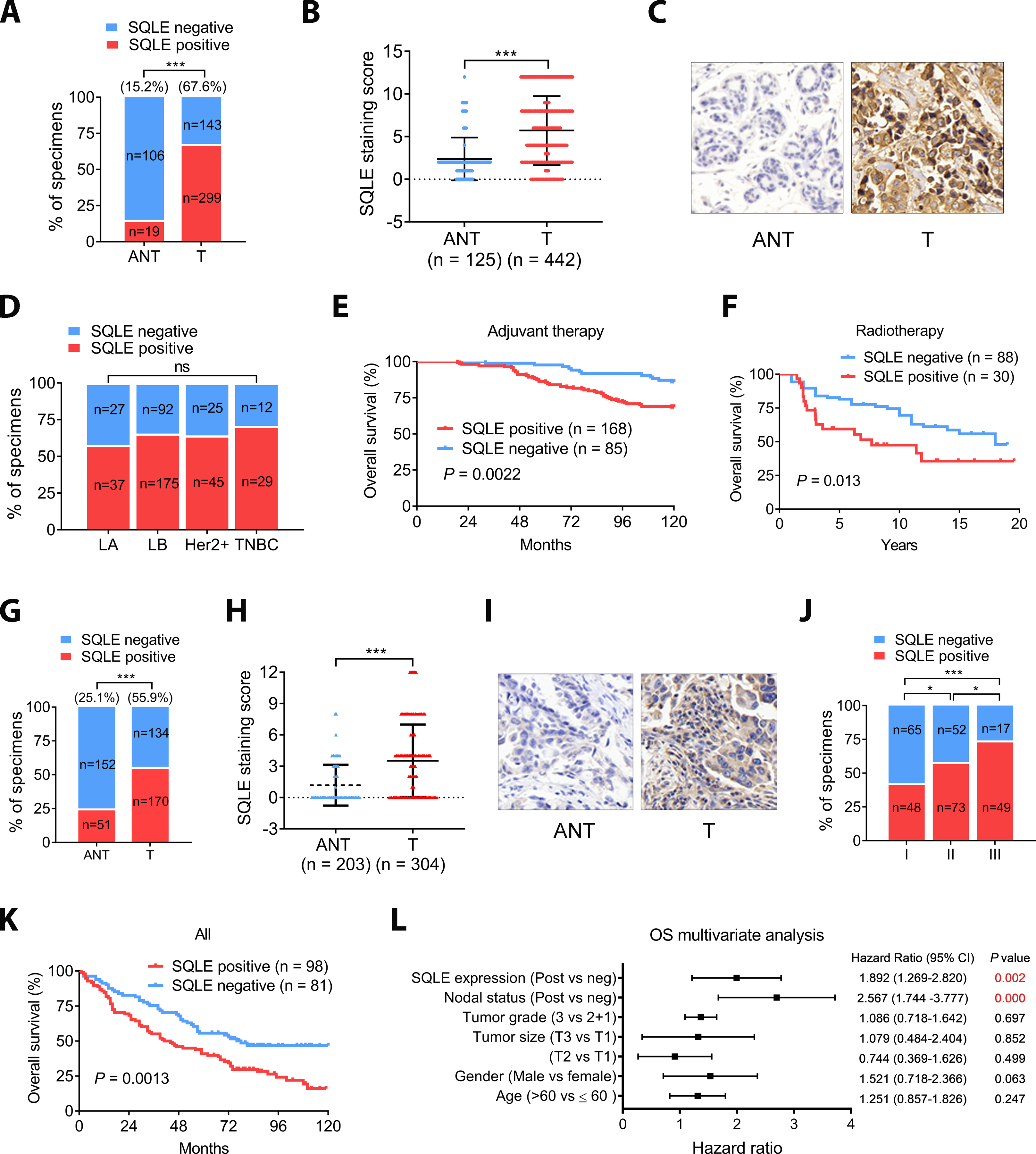
A, SQLE expression was elevated in BC. B, The median SQLE IHC score (Immunoreactive score, IRS) of the BC tumors was significantly higher than that of the ANTs. IRS was determined from the staining intensity (SI) and percentage of positive cells (PP): IRS = SI × PP. An IRS ≥ 3 score was classified as SQLE positive. Groups were compared with the Mann–Whitney test. C, Representative IHC images of matched BC and ANT tissue. Magnification 40×. D, Association of SLQE expression and BC subtypes. E, High SQLE expression correlates with poor survival in a BC population on adjuvant therapy. Kaplan-Meier survival analysis of our cohorts. F, TCGA data analysis showed that high SQLE expression correlates with poor survival in a BC population administered radiation therapy (GSE10374). G, SQLE expression was elevated in NSCLC. SQLE protein expression in NSCLC and ANT was determined using IHC. H, The median SQLE staining of NSCLC tumors was significantly higher than in the ANTs. I, Representative IHC images of NSCLC tumors with more than or less than 3-year survival. Magnification 40×. J, The percentage of positive SQLE staining in NSCLC tumors of different TNM stages. K, High SQLE expression correlated with poor survival in NSCLC. L, Multivariate cox regression analysis of OS in our cohort of lung cancer. Data in (A, D, G and H) was evaluated by chi-square test and in (B and H) was evaluated by Mann-Whitney test. Survival in (E, F and K) were determined using the Kaplan-Meier method, and differences between groups were tested using the log-rank test. Multivariate analysis in (L) was performed with the cox proportional hazard regression model.*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
