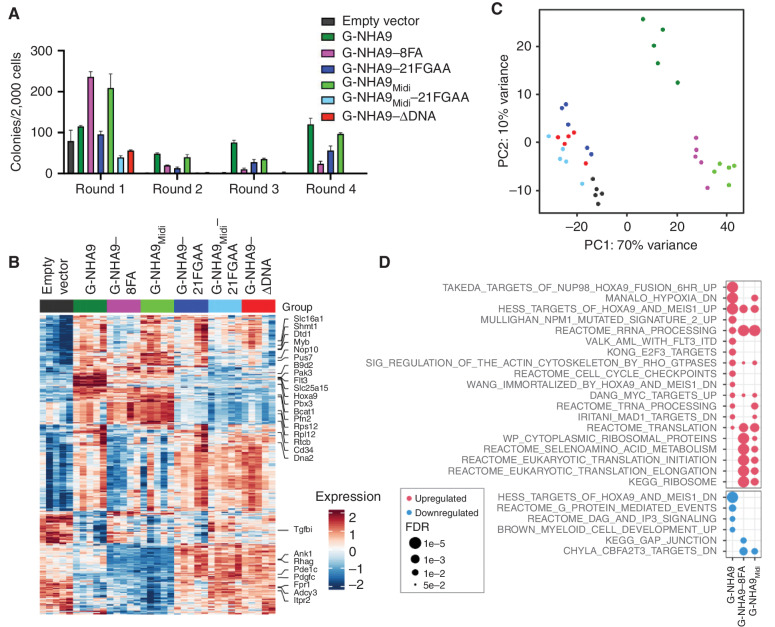
Figure 6.
Expression of high FG motif valence NHA9 constructs in lin− HSPCs leads to hematopoietic cell transformation and aberrant expression of Hox family and other genes. A, Average number of colonies per 2,000 cells for lin− HSPCs expressing negative control empty vector and mEGFP-tagged NHA9 and mutant constructs. The values of colony numbers shown are mean ± SD from triplicate technical replicates of a representative experiment. B–D, RNA-seq was performed for lin− HSPCs expressing empty vector, G-NHA9, or mutants after 1 week of growth in methylcellulose containing myeloid and erythroid growth factors (n = 5 for each condition). B, Heat map for differentially expressed genes of interest. C, PCA of the 500 most variable genes. D, Gene set enrichment analysis for cells expressing G-NHA9, G-NHA9–8FA, or G-NHA9Midi—each versus empty vector. Pathways of interest are shown, with a complete list of significantly upregulated or downregulated gene sets in Supplementary Table S3. The most significantly dysregulated genes from each pathway are marked in B.