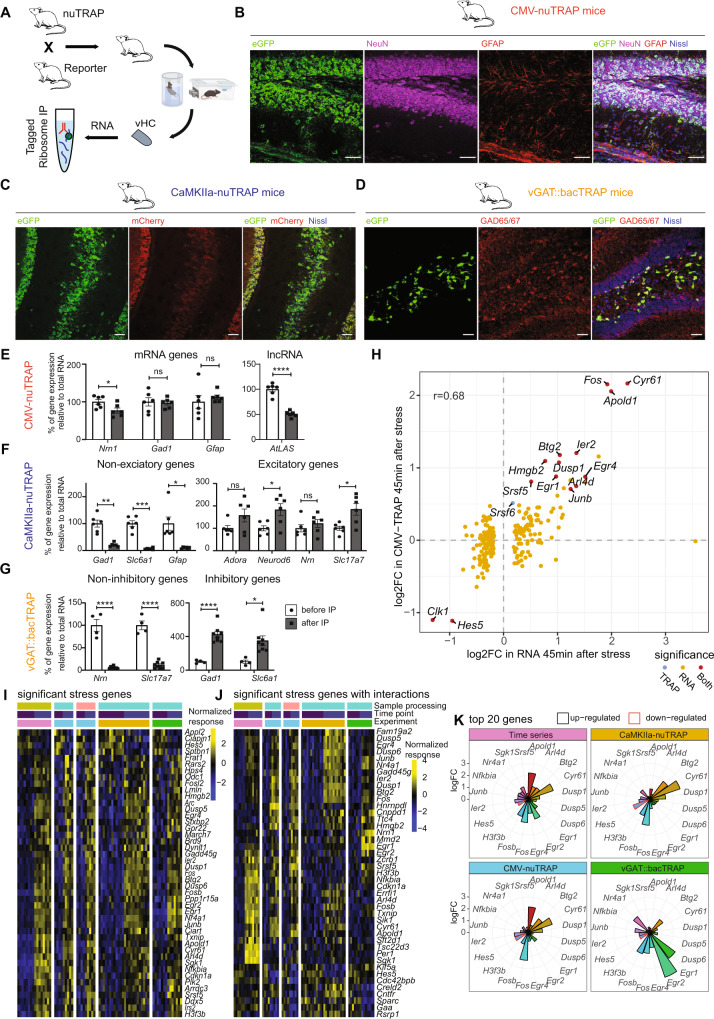
Fig. 6. Acute stress (AS) effects on the bulk translatome, and the translatome of excitatory and inhibitory neurons of the ventral hippocampus (vHC).
A Schematic of TRAP. B-D Microscopy images showing co-localization of eGFP with NeuN and GFAP in the hilus and DG of CMV-nuTRAP mice (B), eGFP and mCherry in the ventral hilus, CA3 and DG of CaMKIIa-nuTRAP mice (C), and eGFP with GAD65/67 in the dorsal hilus and DG of vGAT::bacTRAP mice (D). Scale bars 50 μm. Experiments performed 1-2 times with similar results, images representative of <4 mice/line. E–G RT-qPCR validation of TRAP enrichment. Individual values represent dHC and vHC samples from 3–4 mice/line. Data represent mean ± SEM, asterisks indicate p values generated by independent unpaired t-tests. Source data are provided as a Source Data file (E) Genes primarily expressed in excitatory neurons, inhibitory neurons or glial cells are present both before (total RNA) and after (bound RNA) the IP, while the lncRNA AtLAS is reduced in the bound RNA of hippocampal samples from CMV-nuTRAP mice (Nrn1 t(10) = 2.486, p = 0.0322; Gad1 t(10) = 0.2409, p = 0.8145; Gfap t(10) = 0.683, p = 0.5101, AtLAS t(10) = 9.134, p = 3.62e-06). F Depletion of genes not expresse in excitatory neurons, and enrichment of genes primarily expressed in excitatory neurons after IP compared to before, in hippocampal samples from CamKIIa-nuTRAP mice (Gad1 t(5) = 6.65, p = 0.0012; Slc6a1 t(5) = 9.00, p = 0.0003; Gfap t(5) = 3.69, p = 0.0141; Adora1 t(5) = 2.20, p = 0.0794; Neurod6 t(5) = 3.32, p = 0.0210; Nrn1 t(5) = 1.15, p = 0.3012; Slc17a7 t(5) = 2.87, p = 0.0350). G Depletion of genes not expressed in inhibitory neurons and enrichment of genes primarily expressed in inhibitory neurons after IP compared to before, in hippocampal samples from vGAT::bacTRAP mice (Nrn1 t(10) = 10.49, p = 1.03e-06; Slc17a7 t(10) = 11.67, p = 3.80E-07; Gad1 t(10) = 6.23, p = 9.76E-05; Slc6a1 t(10) = 3.01, p = 0.0131). H Correlation between bulk sequencing and CMV-nuTRAP logFCs 45 mins after stress. Colors indicate significance in analyses (red = significant in both datasets, blue = in CMV-nuTRAP only, yellow = in bulk only). I Significant genes that are AS responsive in pooled data from all experiments (time series, CMV-nuTRAP, vGAT::bacTRAP and CaMKIIa-nuTRAP) in the vHC. J Significant genes when employing a model that includes interaction terms in the vHC. K logFC 45 mins after AS across datasets of the top 20 genes in the vHC. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Male CamKIIa-nuTRAP and vGAT::bacTRAP, and female CMV-nuTRAP were used.