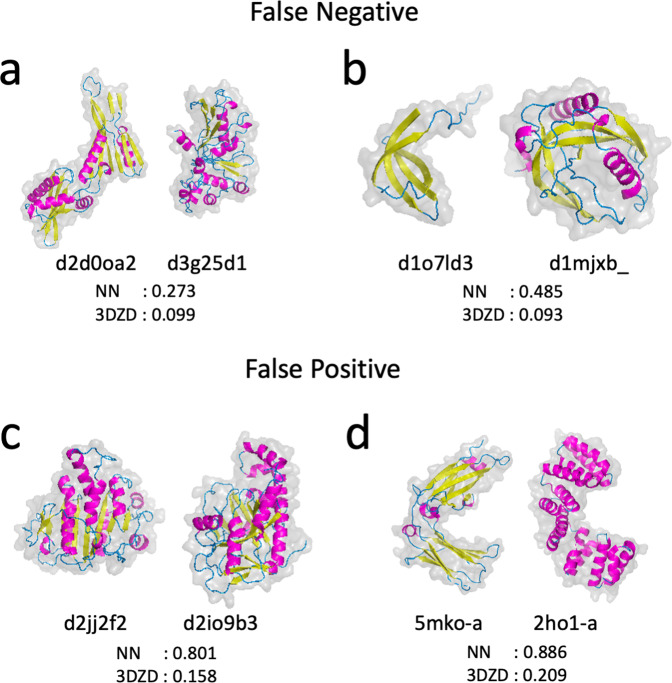
Fig. 4. Examples of protein pairs that were misclassified by 3D-AF-Surfer.
Four protein structure pairs are shown with scores from 3DZD-NN and 3DZD. 3DZDs of the main-chain atoms were used. The two numbers below each protein pair are scores of the two structures by 3DZD-NN and 3DZD. The pairs on panel a and b are cases where both 3DZD-NN and 3DZD considered the two proteins to belong to different SCOP folds but they actually do not. a d2d0oa2 and d3g25d1 belong to the Ribonuclease H-like motif fold (SCOP code: c.55). The scores of 3DZD-NN and 3DZD for this pair was 0.273 and 0.099, respectively, both of which were lower than threshold values used (0.6 and 0.1) and thus considered as different folds. b d1o7ld3 and d1mjxb_ belong to OB fold (b.40). The two pairs on panel c and d are examples of false positives, where 3D-AF-Surfer suggested that each pair belonged to the same fold, but they actually do not. c d2jj2f2 belongs to P-loop containing nucleoside triphosphate hydrolases (c.37) while d2io9b3 belongs to ATP-grasp (d.142). d PDB ID: 5mko-A is a β class structure while 2ho1-A is an α-class structure. These two structures do not have a SCOP ID assigned at time of writing. This example is taken from the paper that reported ZEAL37.