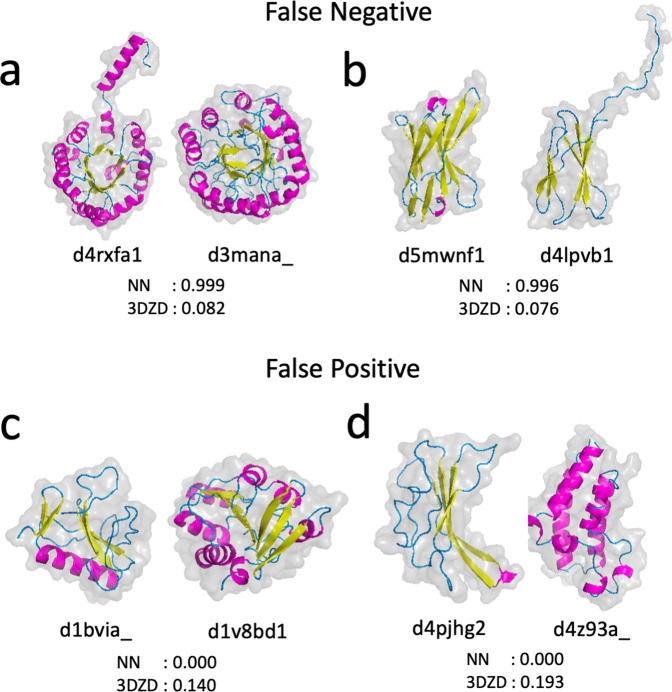
Fig. 5. Examples of pairs where 3DZD-NN classified correctly but 3DZD did not.
Four protein structure pairs with the scores by 3DZD-NN and 3DZD are shown. The main-chain atom representation was used for these comparisons. The two pairs on panel a and b are cases where 3DZD did not recognize them as the same fold (i.e. false negative) but 3DZD-NN did. a d4rxfa1 and d3mana_ belong to the TIM beta/alpha-barrel (SCOP code: c.1). 3DZD-NN had a probability score of 0.99, i.e. very confident that these two structures belong to the same fold while 3DZD had a score of 0.082, below the detection threshold of 0.1. b d5mwnf1 and d4lpvb1, belong to Immunoglobulin-like beta-sandwich (b.1). Panel c and d shows two pairs, where 3DZD-NN correctly detected that the two structures have different folds while 3DZD had a score above the threshold of 0.1 and thus considered them as the same fold. Fold assignment of the four structures in SCOP are as follows: c, d1bvia_: Microbial ribonucleases (d.1); d1v8bd1: NAD(P)-binding Rossmann-fold domains (c.2); (d), d4pjhg2: Immunoglobulin-like beta-sandwich (b.1); d4z93a_: Bromodomain-like (a.29).