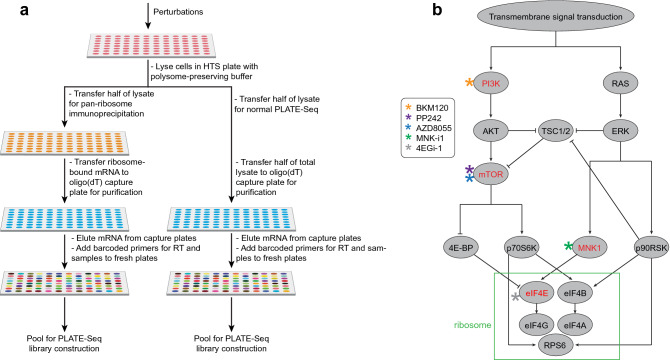
Figure 1.
Overviews of the protocol and experimental design of the study performed. (a) Schematic diagram of the riboPLATE-seq protocol, from lysis in a multi-well plate to pooled library preparation. The right-hand side mirrors the original PLATE-seq protocol. In this workflow, an oligo(dT)-grafted plate captures polyadenylated RNA that can be reverse-transcribed with barcoded adapters, generating a plate of cDNA that may be pooled for library construction. The left side incorporates a pan-ribosome IP before PLATE-seq pooling and library preparation, generating instead a pooled library of ribosome-associated RNA. (b) Simplified structure of the signaling pathways under study and the specific protein targets considered. The PI3K/AKT/mTOR signaling axis at left converges with the MAPK/ERK pathway at right on eIF4E, early in the process of ribosome assembly (green box). The figure also outlines the inhibitors used in this study and their specific targets within these pathways. NVP-BKM120 is a PI3K inhibitor (orange), both AZD8055 and PP242 are mTOR inhibitors (blue and purple, respectively), MNK-i1 is a MNK1 inhibitor (green), and 4EGi-1 is a direct eIF4E inhibitor (black).