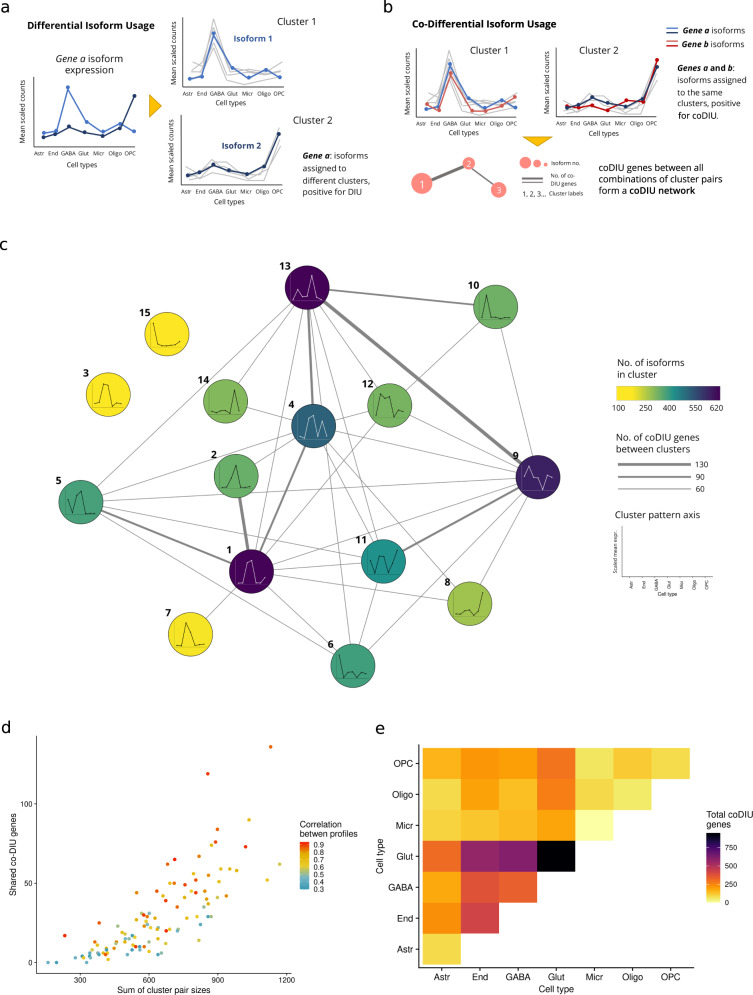
Fig. 4. Characterization of genes with co-Differential isoform usage.
a Cluster-based definition of Differential Isoform Usage (DIU) across multiple cell types. DIU genes have at least two isoforms assigned to different clusters, indicating a differential isoform selection pattern across the different cell types. b Definition of co-Differential Isoform Usage (coDIU) using clusters. CoDIU genes have multiple isoforms assigned to the same clusters, establishinig across-cell type co-expression relationships for at least two of their isoforms. c coDIU network. Nodes represent clusters and depict their mean expression profile across cell types. Node color represents cluster size (i.e., no. of isoforms in cluster). Edge width represents number of coDIU genes detected between each pair of clusters. coDIU genes are considered if they have at least one significant isoform co-expression pattern with one other gene. d Evaluation of cluster profile similarity and size as a function of the number of coDIU genes detected by acorde. X-axis corresponds to the sum of isoforms in each possible pair of clusters generated from the data. Y-axis contains the number of coDIU genes between the pair. Dot color represents the correlation between the mean expression profiles of each pair of clusters. The number of coDIU genes between a pair of clusters is seemingly related to the size of the clusters involved, and shows no relationship with the degree of similarity between the expression profiles of clustered isoforms. e Cell type-level coDIU patterns. For each pair of cell types represented in x and y-axis, heatmap color corresponds to the total number of genes found to be co-DIU between them. Total coDIU genes are calculated as the sum of coDIU genes detected between all cluster pairs that show high expression of isoforms in these cell types. GABA and Glut cell types share the highest number of coDIU genes, both with each other and with other cell types. Astr: astrocytes, End: endothelial cells, GABA: GABA-ergic neurons, Glut: glutamatergic neurons, Micr: microglia, Oligo: oligodendrocytes, OPC: oligodendrocyte precursor cells.