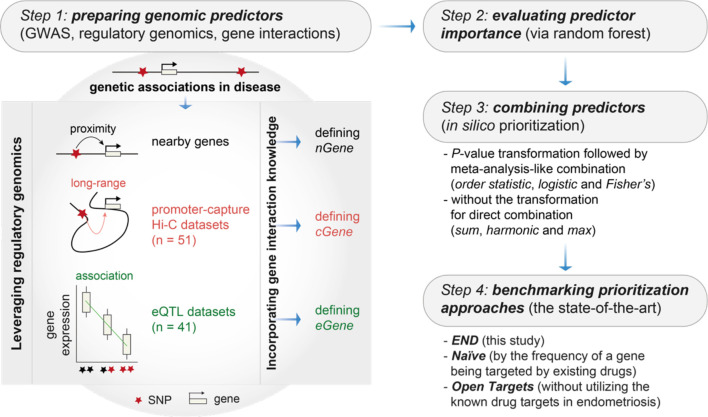
Figure 1.
Overview of genomics-led target prioritization in endometriosis (called ‘END’). Steps 1-3 sequentially describe how to prepare, evaluate and combine genomic predictors for in silico target prioritization. The key is the preparation of genomic predictors at Step 1, as illustrated on the right, utilizing genetic associations in disease, regulatory genomics (mainly promoter-capture Hi-C datasets and eQTL datasets) and the knowledge of gene/protein interactions. Step 4 is intended for benchmarking to compare the performance of END with two competing prioritization approaches, including Naïve (a gene prioritized simply according to how often the gene is already targeted by existing drugs) and Open Targets (without utilizing the information of known drug targets). GWAS, genome-wide association studies; nGene, nearby genes; cGene, conformation genes; eGene, expression genes.