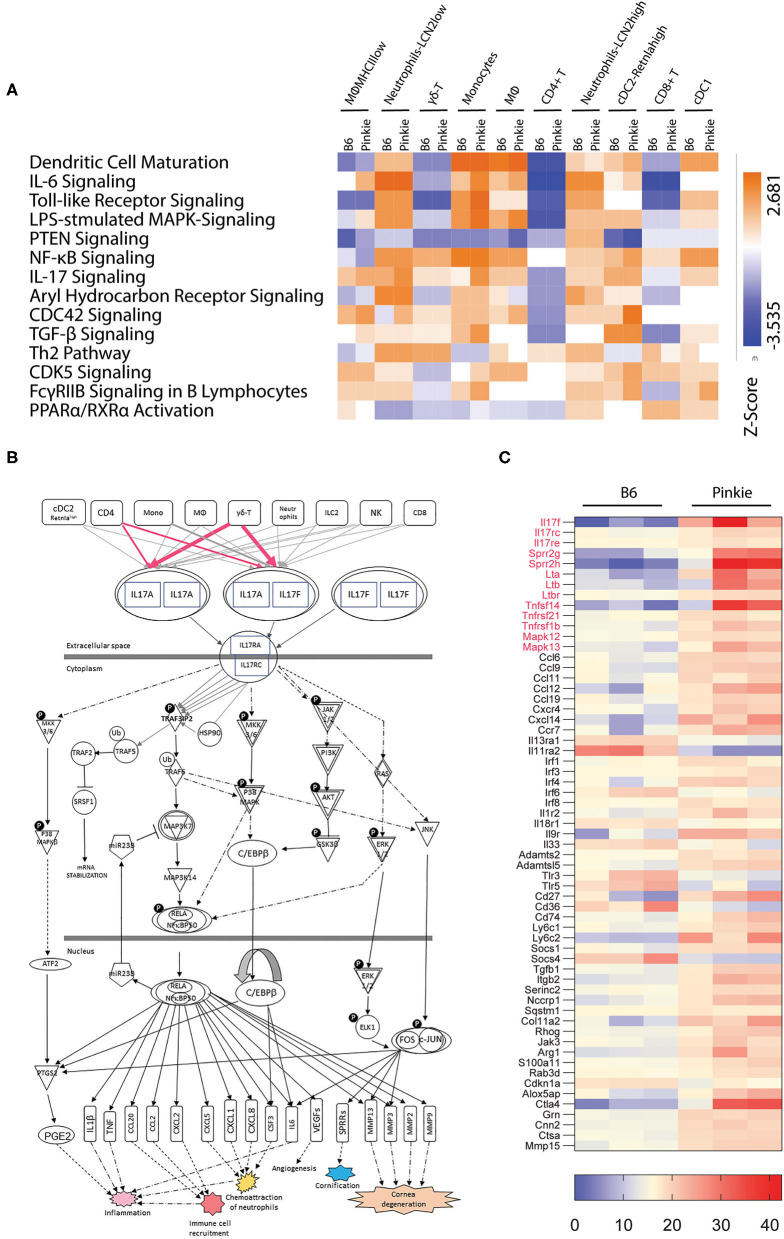
Figure 5.
Pathway analysis. (A) Heatmap of canonical pathways showing significant differences between strains and cell clusters was generated by Qiagen Ingenuity Pathway Analysis. This analysis identified the pathways from the Ingenuity Pathway Analysis library of canonical pathways that were most relevant to the data set. Molecules from the data set that had an adjusted p < 0.05 and were associated with a canonical pathway in the Ingenuity Knowledge Base were considered for the analysis. The significance of the association between the data set and the canonical pathway was measured in two ways: (1) A ratio of the number of molecules from the data set that map to the pathway divided by the total number of molecules that map to the canonical pathway is displayed; and (2) A right-tailed Fisher's Exact Test was used to calculate a p-value determining the probability that the association between the genes in the dataset and the canonical pathway is explained by chance alone. IL-17 signaling and PPARα/RXRα activation pathways are among the pathways identified with significant differences. (B) IL-17 signaling pathway network showing the relationship between molecules generated with Qiagen Ingenuity Pathway Analysis with modification. All connections are supported by at least one reference from the literature, from a textbook, or from canonical information stored in the Ingenuity Knowledge Base. Lines and arrows between nodes represent direct (solid) or indirect (dashed) interactions between gene products and are displayed by cellular localization (extracellular space, plasma membrane, cytoplasm, or nucleus). Rectangles are cytokines and cytokine receptors, triangles are phosphatases, concentric circles are groups or complexes, diamonds are enzymes and ovals are transcriptional regulators or modulators. P, phosphorylation; U, ubiquitination. (C) Heatmap of differentially expressed genes in conjunctival bulk RNA seq between C57BL/6 (B6) and Pinkie strains that includes IL-17 pathway associated genes in red and other innate inflammatory mediators. All the genes passed through the Benjamini-Hochberg procedure to exclude false discovery; the selected genes had an adjusted p > 0.05. Each row represents a specific gene, the right column represents the Pinkie strain and left column represents B6.