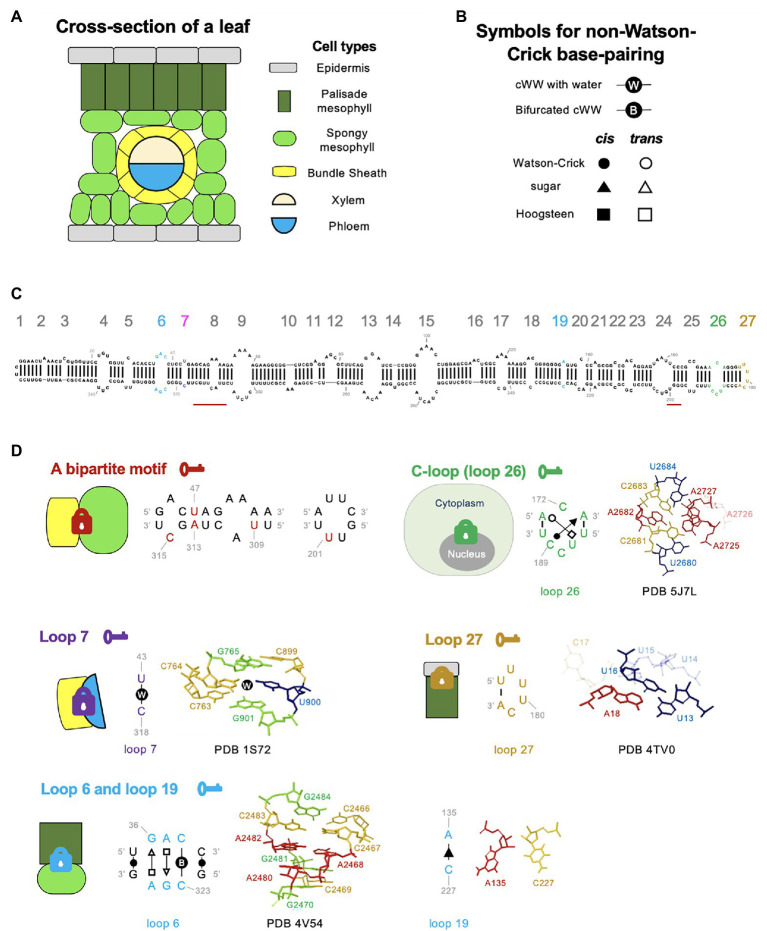
Figure 2.
RNA 3D motif-mediated viroid RNA trafficking in plants. (A) A cartoon illustration of the cross-section of plant leaves. (B) Symbols for annotating RNA nucleotide edges. cWW, cis-Watson Crick-Watson Crick base-pairing. (C) The secondary structure of the PSTVdInt genome, drawing by using RNA2Drawer (Johnson et al., 2019). Loops and bulges are numbered from left to right as 1 to 27. Key structural motifs illustrated in (D) are highlighted in colors (nucleotides and loop numbers). The nucleotides of the bipartite motif (position indicated by two red bars) are not color-highlighted in the secondary structure because this motif was only found in the PSTVdNB strain, whose secondary structure has not been confirmed by chemical probing. (D) Loop structures critical for regulating trafficking across cellular boundaries. Locks with different colors indicate distinct barriers that restrain the trafficking of RNAs without necessary RNA motifs (“keys”). The annotated base-pairings were validated by chemical mapping and functional mutagenesis. The C-loop structure model is based on regions 2,680–2,684 (5′-UCACU-3′) and 2,725–2,727 (5′-AAA-3′) of bacterial 23S rRNA (PDB 5J7L; Cocozaki et al., 2016). The loop 7 structural model is based on regions 763–765 (5′-CCG-3′) and 899–901 (5′-CUG-3′) of Haloarcula marismortui 23S rRNA (PDB 1S72; Klein et al., 2004). The loop 27 model is based on the region 13–18 (5′-UUUUCA-3′) of a Drosophila histone mRNA (PDB 4TV0; Zhang et al., 2014). The loop 6 model is based on regions 2,466–2,470 (5′-CCACG-3′) and 2,480–2,484 (5′-AGACG-3′) of bacterial 23S rRNA (PDB 4V54; Borovinskaya et al., 2007). Note that C2483 and C2467 form a bifurcated cWW base-pairing. The illustration for loop 19 is extracted from the RNA Basepair Catalog. Nucleotides in transparent indicate that they are not involved in base interactions (e.g., A2726 in PDB 5J7L).