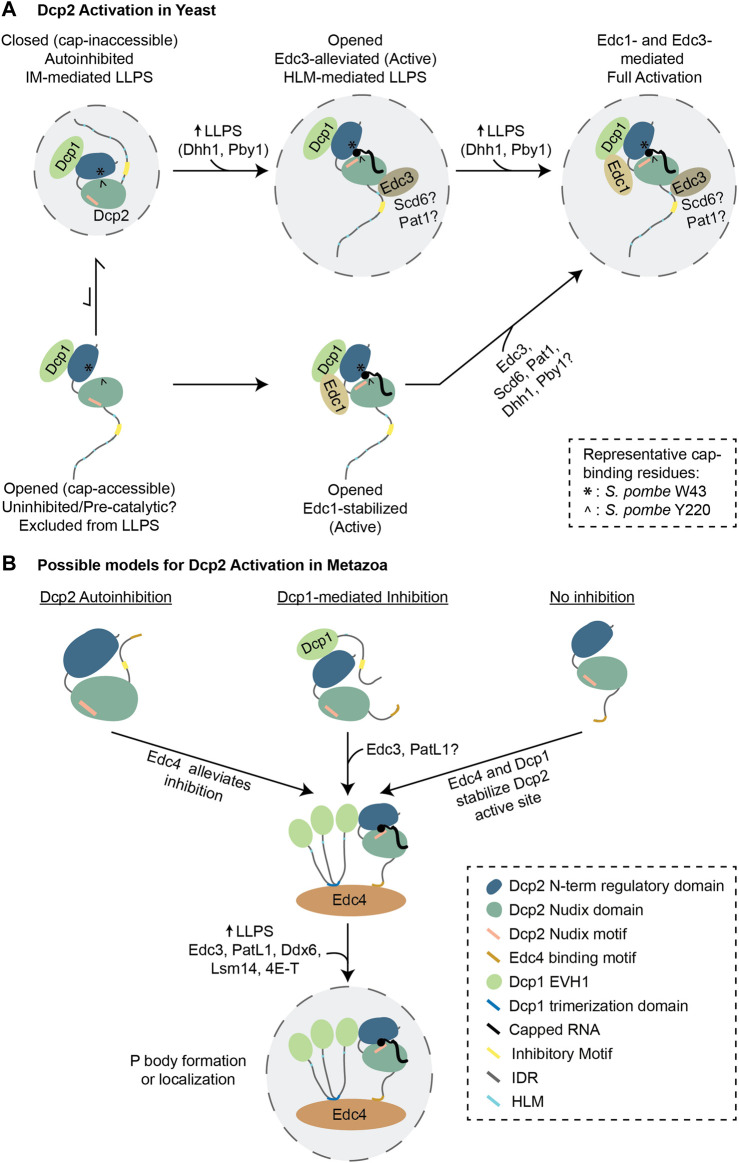
FIGURE 4.
Models of decapping activation in yeast and metazoans. (A) The current model of Dcp2 activation from S. pombe is depicted. Dcp1/2 predominantly exists in an autoinhibited state that self-assemble into LLPS condensates and maintained by the interaction between W49 and Y220. Edc3 binding to Dcp2 HLM likely reorganizes the C-terminal IDR, allowing the formation of an active site in which W49 and Y220 interact with the cap, and activating Dcp2 inside LLPS condensates. On its own, Edc1 may stabilize the opened/active conformation of Dcp2 or consolidate the formation of active site from a pre-catalytic conformation from outside of LLPS condensates. Edc1 can also stabilize the Edc3-alleviated conformation in LLPS condensates, contributing to full activation of Dcp2. (B) Hypothetical model of metazoan Dcp2 activation. It is currently unknown whether or not metazoan Dcp1/2 is regulated through autoinhibition. Since metazoan Edc4 promotes the interaction between metazoan Dcp1 and Dcp2, it may help to alleviate autoinhibition or promote the formation of active site on Dcp2. Other decapping activators might enable decapping by promoting phase separation and Dcp2 localization to LLPS condensates. Abbreviations: IDR = Intrinsically Disordered Region; HLM = Helical Leucine-rich Motif.