Fig. 1: Balancing accessibility, utility, and discovery in clinical genomics.
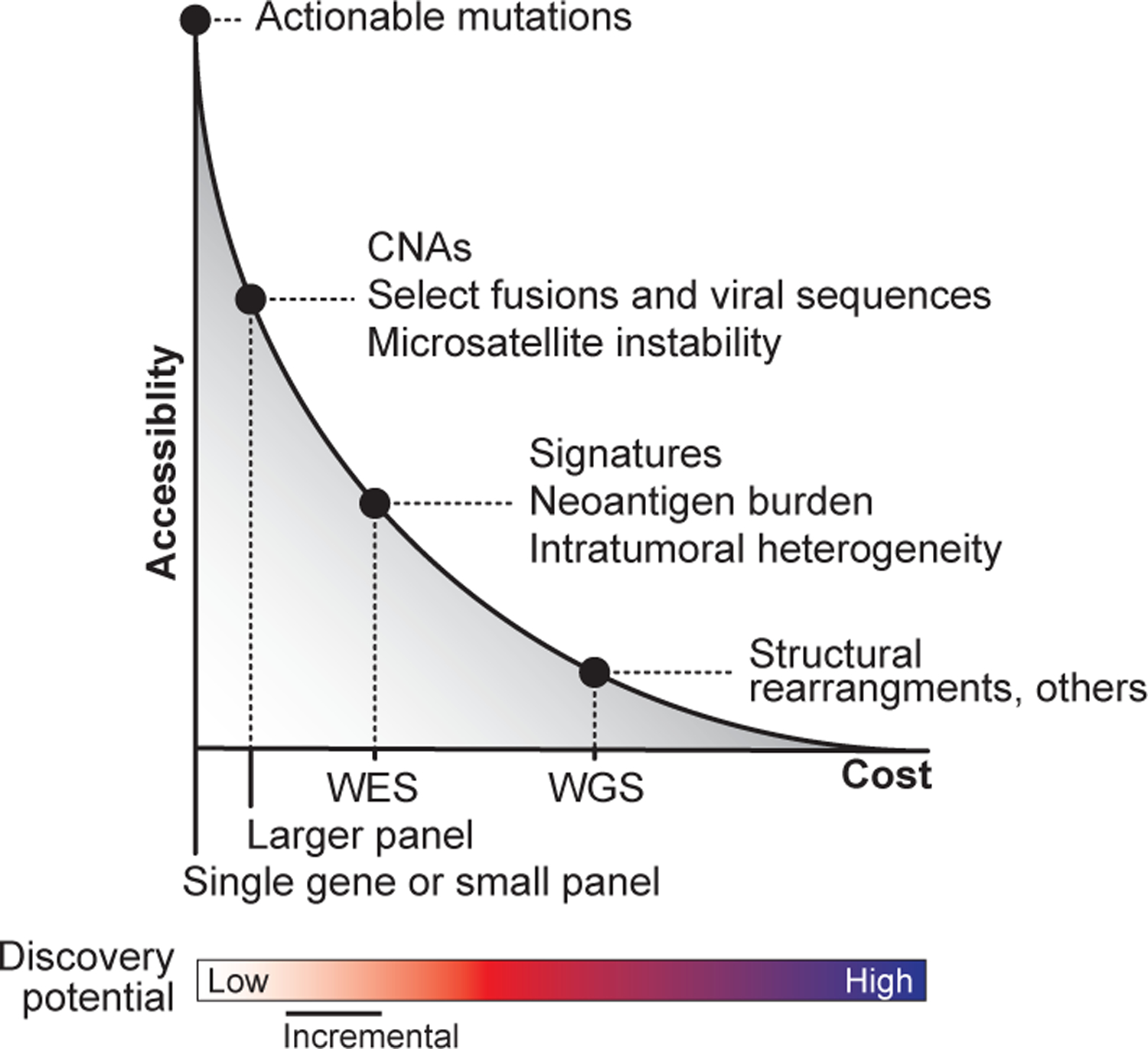
At present, there is a trade-off between cost, accessibility, and the opportunity for novel discovery afforded by different clinical sequencing platforms. The model depicts the current inverse relationship between accessibility and cost. As the cost (x-axis) of different sequencing strategies increases (driven by the size of the genomic footprint and depth of sequencing) accessibility in the community decreases (y-axis). The most common modalities of sequencing are labeled as are examples of the additional types of information enabled possible by adopting the indicated sequencing strategy. The discovery potential of a given strategy typically tracks with cost (bottom). At one end of the spectrum, small gene panels have the lowest cost but have limited discovery potential. In contrast, at the other end of the spectrum, WGS has both the highest cost and greatest discovery potential. WES, whole-exome sequencing; WGS, whole-genome sequencing; CNAs, copy number alterations.
