Figure 1. Overview of iNDI.
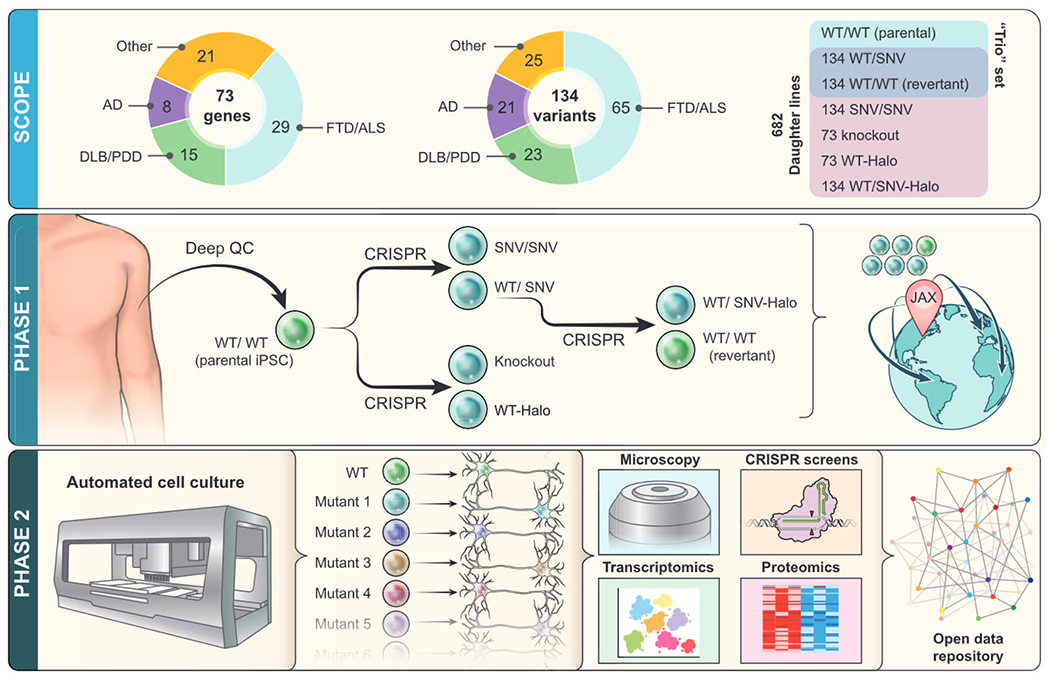
iNDI is a large iPSC genome-engineering initiative, with more than 600 engineered daughter lines to be generated from each parental line. Companion lines will include revertants, Halo-tagged knockins, and knockouts. Phase 1: iNDI will genetically engineer isogenic iPSC lines harboring many ADRD variants, knockouts, and endogenous tags across several deeply characterized parental lines from unaffected individuals. Following stringent quality control assays, each line will be available for distribution through The Jackson Laboratory (JAX). Phase 2: using high-throughput robotic platforms, iNDI will differentiate mutation harboring iPSC lines into CNS-relevant cell types, such as neurons and microglia, followed by a series of phenotypic assays. These include transcriptomics, proteomics, imaging, and genetic interaction screens, yielding a rich multi-dimensional picture of how mutations disrupt cellular function. Open access datasets will enable the community to analyze these data in new ways.
