Figure 1. Overview of nascent matrix labelling approach.
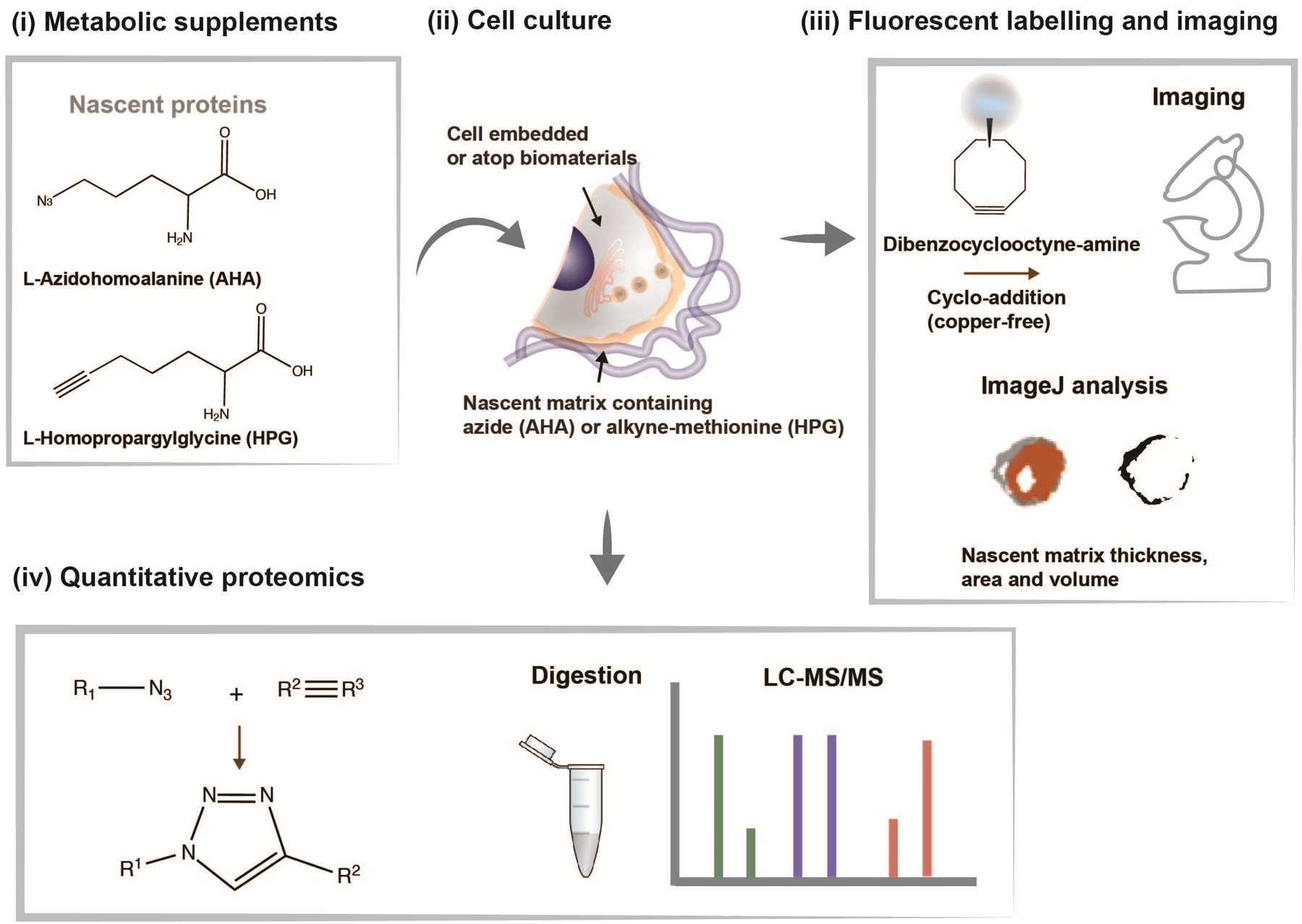
(i) Chemical structures of analogs of the amino acid methionine (L-azidohomoalanine (AHA) and L-homopropargylglycine (HPG)) to label nascent proteins. (ii) Schematic of cell interacting with a biomaterial and cultured in methionine-free media supplemented with AHA or HPG to label nascent proteins. (iii) Incorporated methionine analogs can be fluorescently labelled using click-chemistry (copper-free cycloaddition) using a fluorophore-conjugated cyclooctyne (e.g., dibenzocyclooctyne-amine (DBCO)), imaged and nascent matrix properties (thickness, area, volume) quantified using ImageJ. (iv) Incorporated methionine-analogs can be quantified using liquid chromatography-mass spectrometry (LC-MS/MS) upon release from the biomaterial.
