Fig. 3.
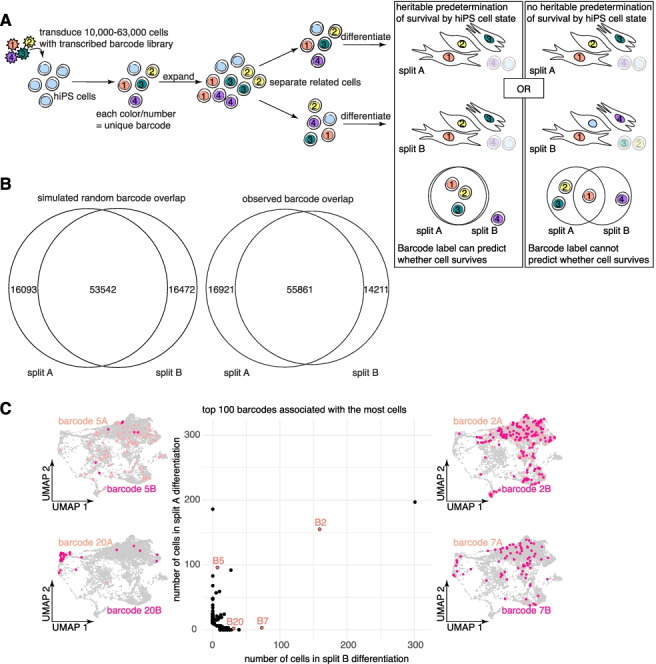
Cellular survival during cardiac directed differentiation is not dictated by differences in hiPS cells. a Schematic representation of experimental workflow. Briefly, we transduced hiPS cells at a range of MOIs (0.1, 0.23, 0.5, or 1.0), then 3 days later (3–4 population doublings), we harvested a third of cells for immediate extraction of genomic DNA and split the remaining cells across two parallel cardiac differentiations. On day 15 of differentiation, we harvested differentiated cells from both splits for genomic DNA extraction. We sequenced and recovered barcodes from the genomic DNA from each split, asking whether the number of overlapping barcodes between splits was greater than would be found by random chance (which would suggest that barcoded clones are predisposed to survival vs death). b Comparison of the simulated random barcode overlap (middle number) across splits with the observed barcode overlap from the splits differentiated from hiPS cells transduced at an MOI of ~0.5 (the experimental condition with the highest observed barcode overlap). c Scatter plot showing the number of cells per split labeled by the top 100 barcode clones composed of the most cells. Maintaining the organization provided by UMAP, we plotted all barcoded cells (gray) and recolored cells corresponding to each of four featured barcodes (marked in red on the scatter plot) in two shades of pink corresponding to the two parallel splits. There is little concordance for most barcode clones between the number of cells that are recovered from each split following cardiac differentiation
