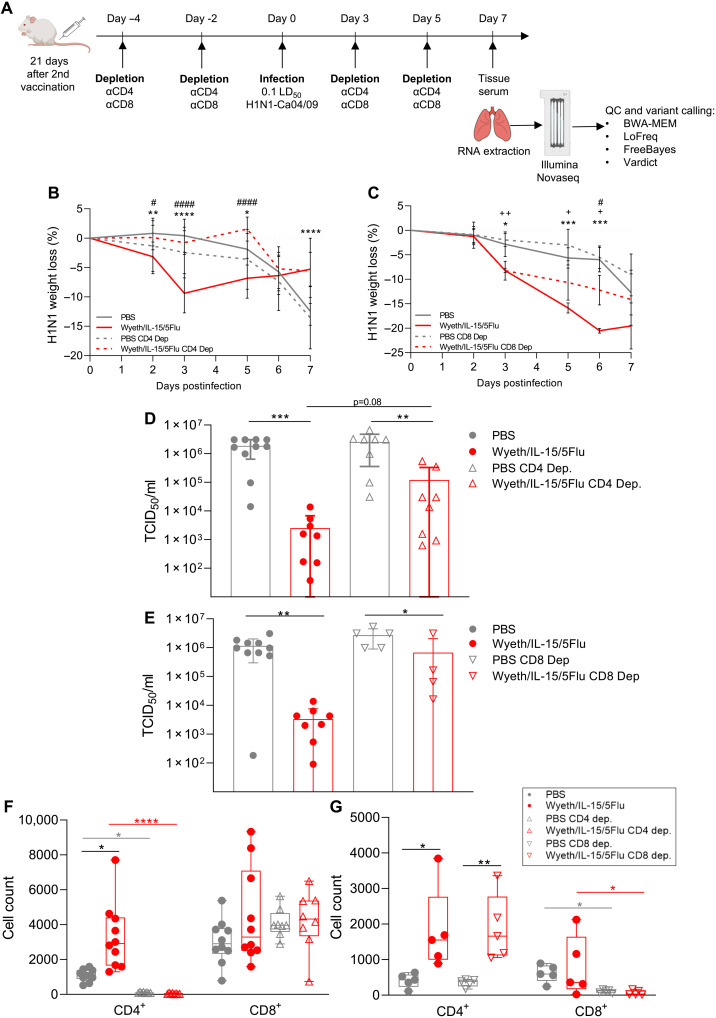
Fig. 3. T cell depletion of BALB/c mice and subsequent challenge with H1N1.
(A) T cell depletion schedule of PBS- or Wyeth/IL-15–vaccinated mice. Infected lungs were harvested at day 7 postinfection, and IAV RNA was extracted, amplified by RT-PCR, sequenced by NovaSeq, and analyzed. Weight loss during H1N1 infection was monitored in (B) PBS and Wyeth/IL-15/5Flu nondepleted (n = 10 ± SD) and CD4+ T cell depleted (n = 8 ± SD). (C) PBS and Wyeth/IL-15/5Flu nondepleted (n = 10 ± SD) and CD8+ T cell depleted (n = 5 ± SD). Significance was determined by two-way ANOVA (* denotes comparison between nondepleted PBS versus Wyeth/IL-15/5Flu, # denotes nondepleted Wyeth/IL-15/5Flu versus T cell–depleted Wyeth/IL-15/5Flu, and + denotes T cell–depleted PBS versus Wyeth/IL-15/5Flu). Lung viral loads (±SD) by TCID50 assay comparison of nondepleted PBS and Wyeth/IL-15/5Flu versus (D) CD4+ T cell–depleted and (E) CD8+ T cell–depleted PBS and Wyeth/IL-15/5Flu–vaccinated groups. H1N1-specific IFN-γ+ CD4+ CD8+ T cell populations from BAL fluid at day 7 p.i. in (F) CD4+ and (G) CD8+ T cell–depleted conditions. Individual data points shown on box and whiskers plot, showing mean and upper and lower quartiles with minimum-maximum range. Statistical significance was determined by a Kruskal-Wallis test. False discovery rate (0.1) was determined by the original Benjamini and Hochberg method. *P < 0.05, **P < 0.01,***P < 0.005, and ****P < 0.0001.