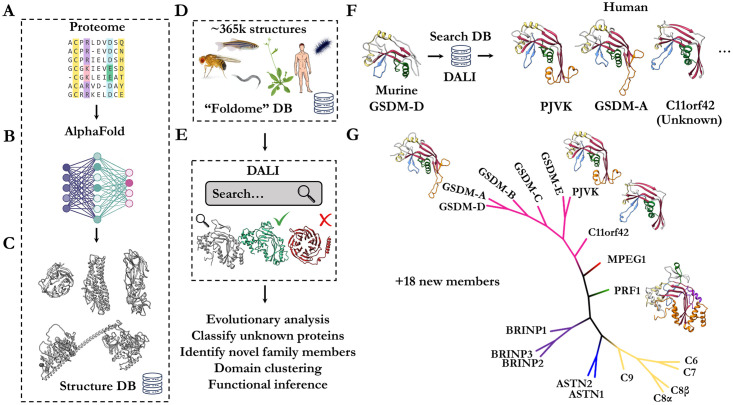
Fig 1. Conceptual overview of structure-guided fold recognition against the AlphaFold database.
a. Entire proteome sequence databases are converted by (b) machine-learning methods into high-accuracy (c) structural model predictions. d. Curation of these structural databases into a unified resource of structures (S1 Text). e. DALI based fold matching to perform functional inference, identification of unknown members and structural classification. f. Searching the foldome in (d) for matches of murine GSDM-D N-terminal domain yields MACPF/GSMD family members, including C11orf42 –an unknown member of the GSDM family. g. Example phylogenetic analysis of perforin-like proteins identified from the foldome by DALI. Alignment of the central MACPF/GSDM fold is possible by extracting domain boundaries based on AlphaFold prediction allowing specific comparison between members without interfering ancillary domains.