Figure 3.
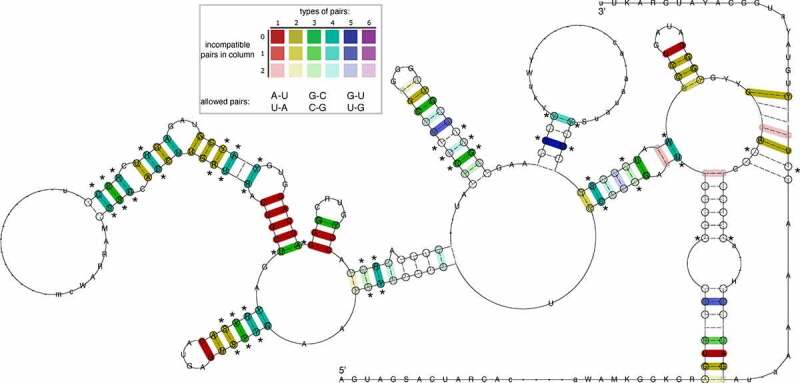
The consensus of IRES Domain III secondary structure prediction generated for 13 Pestivirus genomes highlights rich covariation patterns by the different colouring of base pairs, following the RNAalifold colour scheme. Red indicates nucleotide-level sequence conservation, while other colours highlight increasing covariation levels by structure-conserving nucleotide substitutes, as depicted in the insert. The model includes nucleotides with a frequency greater than the average in the underlying alignment in IUPAC notation. Dashes along the consensus sequence indicate positions where the majority of sequences had gaps. Circled nucleotides highlight compensatory mutations, i.e. cases where both nucleotides of a base pair are mutated, such as UA -> GC. Statistical significantly covarying base pairs are marked with an asterisk.
