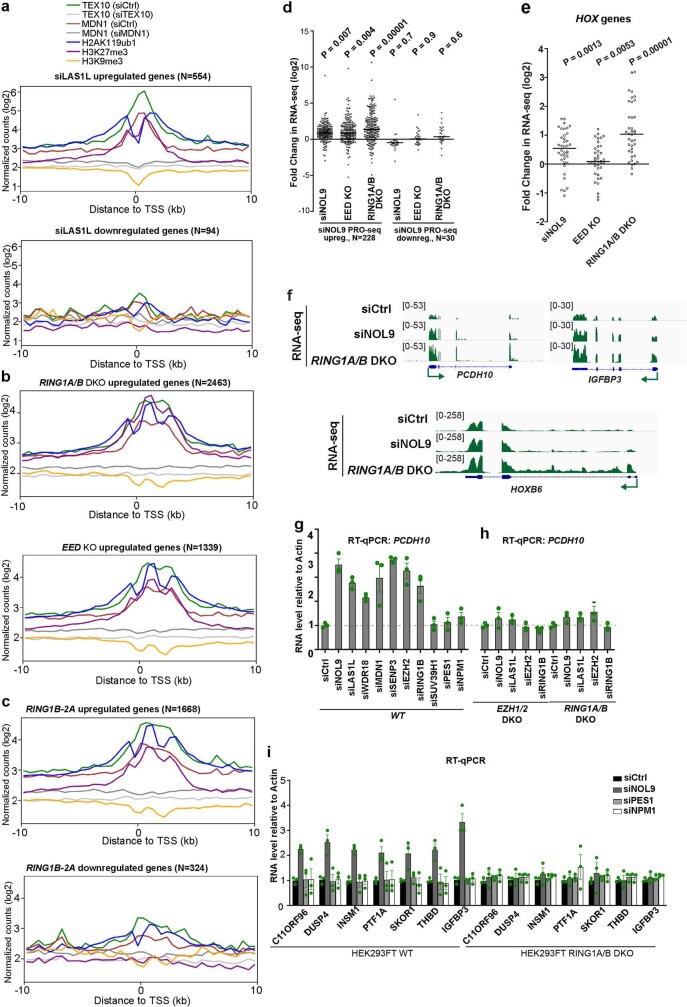
Extended Data Fig. 7. Coregulation of target genes by the rixosome and PRCs.
a–c, Average distribution of the indicated ChIP-seq reads (log2) for genes upregulated and downregulated in LAS1L KD (a) and RING1B-2A(b), and upregulated genes in EED KO and RING1A/B DKO (c) RNA-seq experiments from HEK293FT cells. Enrichment levels were normalized with Reads Per Genome Coverage. Read counts per gene were summed in 50-nt bins. d, Dot plots showing RNA-seq changes in the expression of siNOL9-upregulated or siNOL9-downregulated genes in PRO-seq experiments in HEK293FT cells. siNOL9 PRO-seq upregulated genes have increased RNA levels in siNOL9, EED KO, and RING1A/B DKO cells. p value is from the two-tailed Wilcoxon test. The measure of center is median. e, RNA-seq experiments showing increased HOX gene expression in siNOL9 and RING1A/B DKO but not EED KO cells. P value is from the two-tailed Wilcoxon test. The measure of center is median. f, Genomic snapshots of RNA-seq reads showing the effect of siRNA knockdown of NOL9 and RING1A/B DKO on the expression of the indicated genes in HEK293FT cells. g, h, RT-qPCR assays showing that siRNA knockdown of rixosome subunits results in increased expression of PCDH10 in wild-type (WT), but not EZH1/2 DKO or RING1A/B DKO HEK293FT cells. Actin (ACTB) served as a normalization control. Every knockdown was normalized to siCtrl. Nucleolar PES1 and NPM1 served as controls for possible non-specific effects resulting nucleolar perturbations. Error bars represent standard deviations for three biological replicates. Data are presented as mean values +/− SEM. i, RT-qPCR experiments showing the effect of the indicated siRNA knockdowns on the indicated Polycomb and rixosome target genes in wild-type (WT) cells and RING1A/B DKO cells. Actin (ACTB) served as a normalization control. Every knockdown was normalized to siCtrl. Error bars represent standard deviations for three biological replicates. Data are presented as mean values +/− SEM.