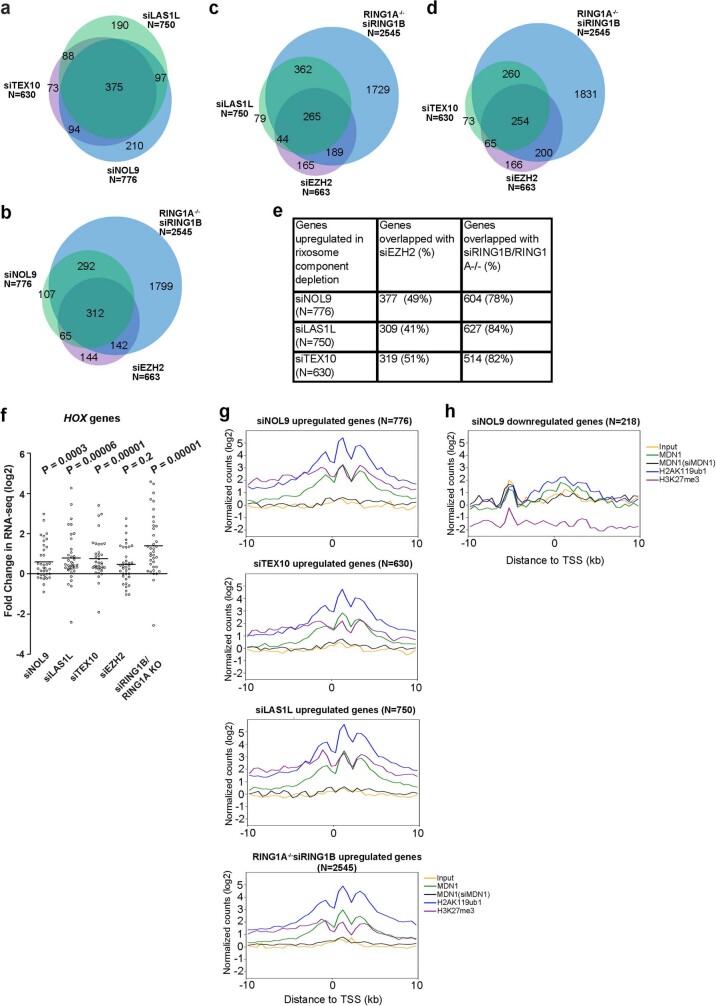
Extended Data Fig. 9. Role of the rixosome in Polycomb silencing is not cell type specific.
a, Venn diagram showing overlap among genes upregulated in RNA-seq analysis of siNOL9, siLAS1L, and siTEX10 HeLa cells. Hypergeometric probability p values: siNOL9 vs siLAS1L, 2.6e-765; siNOL9 vs siTEX10, 1.4e-859; siLAS1L vs TsiEX10, 1.6e-853. b, Same as in a but showing overlap among genes upregulated in siRING1B in RING1A KO (siRING1B, RING1A-/-), siNOL9 (in wild type), and siEZH2 (in wild type). Hypergeometric probability p values: siNOL9 vs siRING1B, RING1A-/-, 4.1e-441; siNOL9 vs siEZH2, 4.6e-411; siEZH2 vs siRING1B, RING1A-/-, 2.9e-284. c, Same as in a but showing overlap among genes upregulated in siRING1B, RING1A-/-, siLAS1L (in wild type), and siEZH2 (in wild type). Hypergeometric probability p values: siLAS1L vs siRING1B, RING1A-/-, 1.4-496; siLAS1L vs siEZH2, 1.7e-298. d, Same as in a but showing overlap among genes upregulated in siRING1B, RING1A-/-, siTEX10 (in wild type), and siEZH2 (in wild type). Hypergeometric probability p values: siTEX10 vs siRING1B, RING1A-/-, 1.8e-391; siTEX10 vs siEZH2, 9.1e-347. e, Table showing the percentages of overlapping upregulated genes between rixosome and PRC depletions in panels a–d. f, Dot plots of RNA-seq experiments showing changes in the expression of 39 HOX genes in HeLa cells. p values are from two-tailed Wilcoxon test. The measure of center is median. g, Average distribution of indicated ChIP-seq reads (log2) for genes upregulated by siRNA depletion of TEX10, LAS1L, NOL9, and RING1B (RING1A-/-) in HeLa cell RNA-seq experiments. Enrichment levels were normalized with Reads Per Genome Coverage. Read counts per gene were summed in 50-nt bins. h, Same as in g but showing siNOL9 downregulated genes.