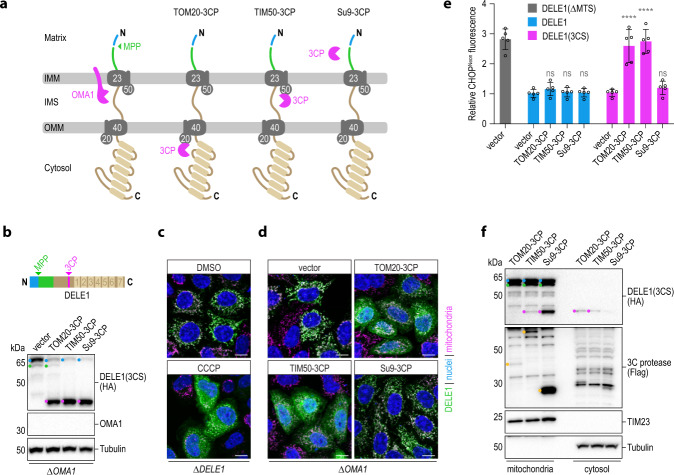
Fig. 4. Cleavage of DELE1 in the IMS is sufficient for its release to the cytosol and activation of the ISR.
a Schematic depicting the localization of the proteases employed in this figure relative to the DELE1 protein during its import into mitochondria. b HeLa OMA1 knockout (ΔOMA1) cells were transiently transfected with DELE1(3CS) and empty vector control or the indicated 3C protease fused to TOM20, TIM50 or the sorting signal of Su9. Cleavage of the DELE1 protein was analyzed by immunoblotting. c, d HeLa cells were transiently transfected with DELE1(3CS) as in (b). Localization of the DELE1 protein was analyzed by confocal microscopy after a 2 h treatment with DMSO or CCCP (c) or in the context of the co-transfected 3C proteases (d). Scale bars, 10 μm. Nuclei (DAPI, blue), mitochondria (MitoTrackerRed, pink), DELE1 (HA, green). e The induction of the ISR marker CHOP was measured in HAP1 CHOPNeon OMA1 knockout cells by flow cytometry upon transient transfection of the indicated constructs together with mCherry. Relative CHOPNeon fluorescence to empty vector transfected cells is shown. Graph depicts mean ± s.d. of n = 5 independent experiments. DELE1(ΔMTS) was used as positive control. Statistical significance within DELE1 constructs compared to the respective vector control was assessed by ordinary one-way ANOVA and Dunnett’s multiple comparisons correction. DELE1: ns ≥ 0.4019; DELE1(3CS): ****P < 0.0001, ns = 0.8269. f HeLa DELE13CS-HA cells were transfected with the indicated Flag-tagged 3C protease (yellow dots) and cleavage and localization of DELE1 species in mitochondria or the cytosol was analyzed upon subcellular fractionation by immunoblotting.