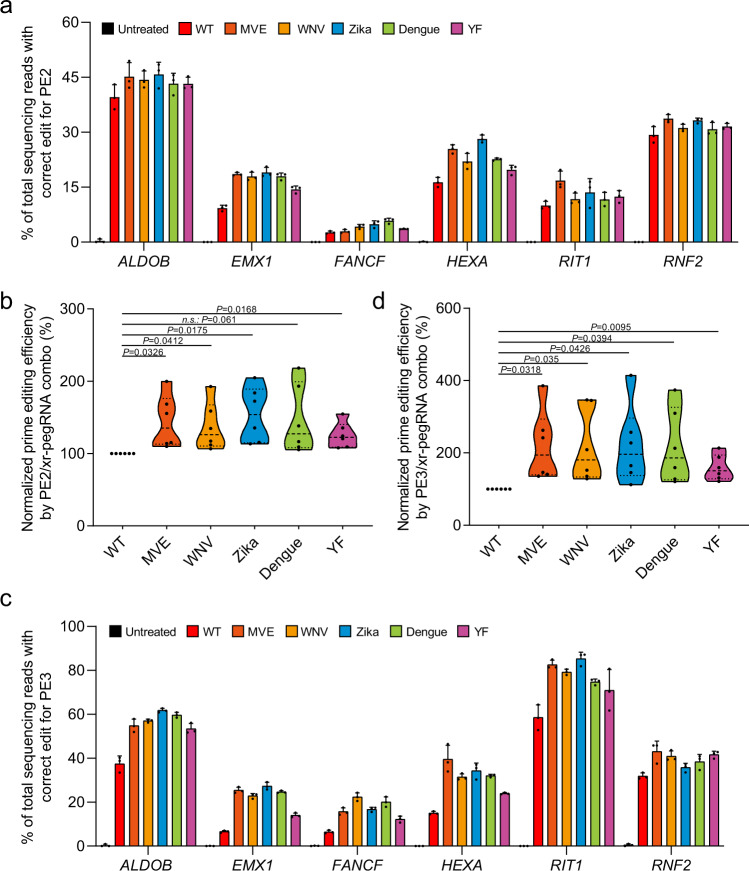
Fig. 2. The xr-pegRNA enhances prime editing of base conversions at various sites within genomic context.
a HEK293T cells were co-transfected with plasmids for PE2 and WT pegRNAs or different xr-pegRNAs targeting indicated (6) genomic loci for base conversions. Following isolation of genomic DNA from transfected cells (sorted), correct editing rates at each site were determined by deep-sequencing (mean ± SD, n = 3 biological replicates). Reads that only contain the intended edits were counted. b. Results in a is further analyzed by considering editing at all sites (n = 6 sites) as a whole. The editing frequencies induced by PE2 with WT pegRNA were set as 100%. c. Experiments were carried out similar to (a), except that a PE3 strategy was used (mean ± SD, n = 3 biological replicates). d Results in c is further analyzed by considering editing at all sites (n = 6 sites) as a whole. The editing frequencies induced by PE3 with WT pegRNA were set as 100%. In the violin plots shown in b and d, each point represents the averaged editing activity at the particular site. The thicker dotted line shows the medians of all data points, while the thinner dotted lines correspond to quartiles (1st and 3rd). Two-tailed one-sample Student’s t tests were performed. The P values are marked on the graphs (n.s. not significant). Source data are provided as a Source Data file.