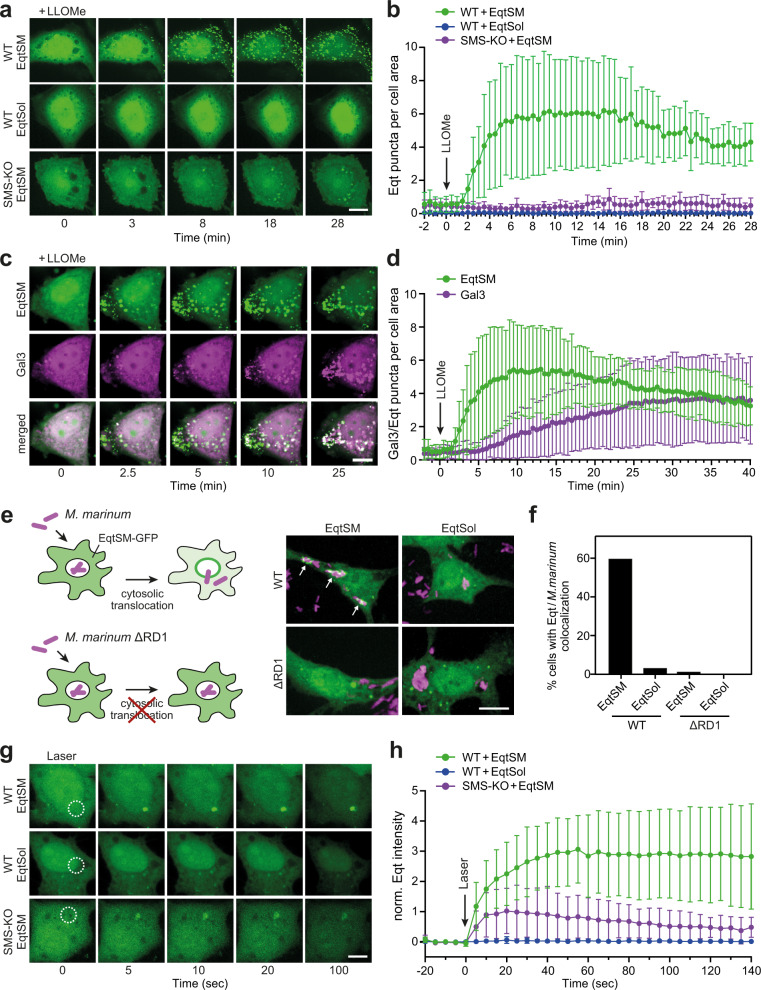
Fig. 1. Cytosolic EqtSM readily binds organelles injured by chemicals, pathogens, or light.
a Time-lapse images of wild-type (WT) or SMS-KO HeLa cells expressing GFP-tagged EqtSM or EqtSol and treated with 1 mM LLOMe for the indicated time. b Time-course plotting Eqt-positive puncta per 100 μm2 cell area in cells treated as in (a). Data are means ± SD. n = 9 cells for WT + EqtSM, 9 cells for WT + EqtSol, 8 cells for SMS-KO + EqtSM over two independent experiments. c Time-lapse images of wild-type HeLa cells co-expressing GFP-tagged EqtSM (green) and mCherry-tagged Gal3 (magenta) treated with 1 mM LLOMe for the indicated time. d Time-course plotting Eqt- and Gal3-positive puncta per 100 μm2 cell area in cells treated as in (c). Data are means ± SD. n = 17 cells. e RAW264.7 cells expressing GFP-tagged EqtSM or EqtSol (green) were infected with mCherry-expressing wild-type (WT) or translocation-defective (ΔRD1) mutant strains of Mycobacteria marinum (magenta). Live-cell fluorescence micrographs were captured 2 h post-infection. f Percentage of cells treated as in (e) showing co-localization of mCherry-expressing M. marinum and EqtSM-positive puncta. n = 30 cells per condition over two independent experiments. g Time-lapse images of wild-type (WT) or SMS-KO HeLa cells expressing GFP-tagged EqtSM or EqtSol and locally wounded by a brief pulse from a 2-photon laser. h Time-course plotting Eqt-associated fluorescence at the laser-induced wound site in cells treated as in (g). Data are means ± SD. n = 7 cells for WT + EqtSM, 4 cells for WT + EqtSol, 6 cells for SMS-KO + EqtSM over two independent experiments. Scale bar, 10 µm.