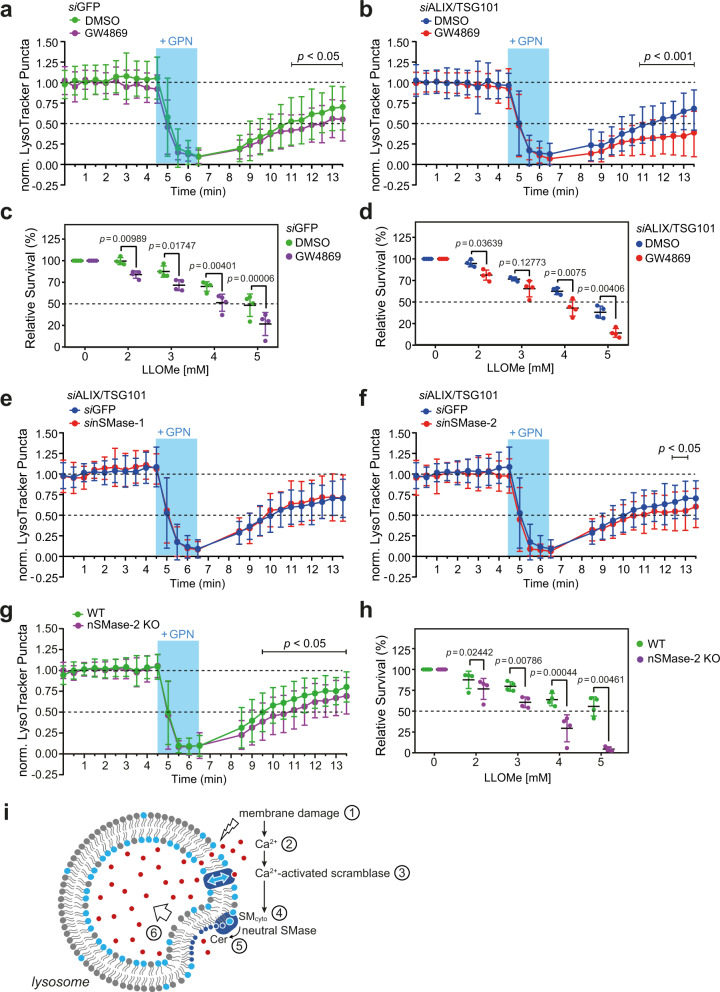
Fig. 7. Inhibition of neutral SMases disrupts repair of damaged lysosomes.
a Time-course plotting LysoTracker-positive puncta in HeLa cells pre-treated with siRNA targeting GFP (72 h) and GW4869 (10 µM) or DMSO (0.5%, 30 min) and pulse-treated with GPN. Data are means ± SD. n = 52 cells for DMSO and 39 cells for GW4869 over three independent experiments. b Time-course plotting LysoTracker-positive puncta in HeLa cells pre-treated with siRNAs targeting ALIX/TSG101 and GW4869 or DMSO and pulse-treated with GPN. Data are means ± SD. n = 46 cells for DMSO and 44 cells for GW4869 over three independent experiments. c Survival rate of HeLa cells pre-treated with siRNA targeting GFP after 5 h of exposure to LLOMe at indicated concentrations in the presence of GW4869 or DMSO. Data are means ± SD. n = 3 independent experiments. d Survival rate of HeLa cells pre-treated with siRNA targeting ALIX/TSG101 after 5 h exposure to LLOMe at indicated concentrations in the presence of GW4869 or DMSO. Data are means ± SD. n = 3 independent experiments. e Time-course plotting LysoTracker-positive puncta in HeLa cells pre-treated with siRNAs targeting ALIX/TSG101 and GFP or nSMase-1 and pulse-treated with GPN. Data are means ± SD. n = 28 cells for siGFP and 28 cells for sinSMase-1 over three independent experiments. f Time-course plotting LysoTracker-positive puncta in HeLa cells pre-treated with siRNAs targeting ALIX/TSG101 and GFP or nSMase-2 and pulse-treated with GPN. Data are means ± SD. n = 28 cells for siGFP and 40 cells for sinSMase-2 over three independent experiments. g Time-course plotting LysoTracker-positive puncta in WT or nSMase-2 KO HeLa cells pulse-treated with GPN. Data are means ± SD. n = 58 cells for WT and 66 cells for nSMase-2 KO over three independent experiments. h Survival rate of wild-type (WT) or nSMase-2 KO HeLa cells after 5 h exposure to LLOMe at the indicated concentration. Data are means ± SD. n = 4 independent experiments. P values were calculated by paired (c, d, h) or unpaired two-tailed t test (a, b, e, f, g). i Model illustrating how Ca2+-activated SM scrambling and turnover may promote restoration of damaged lysosomes.