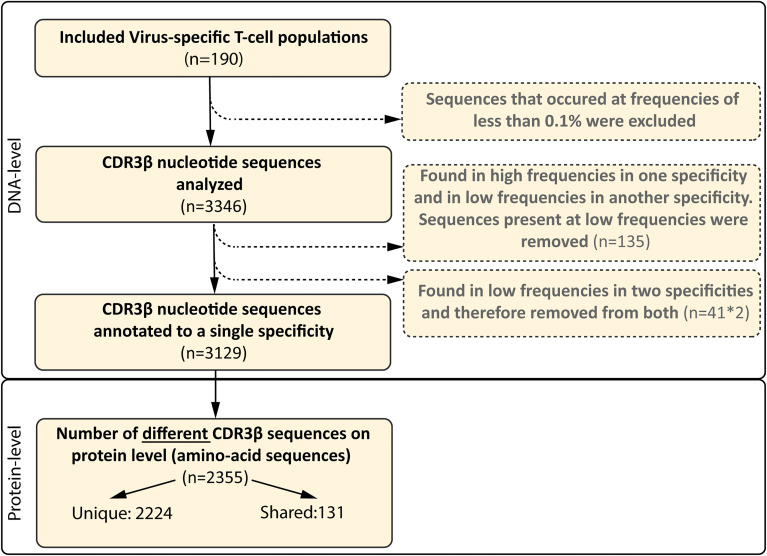
Figure 1.
Flowchart of included and excluded CDR3β nucleotide and AA-sequences. In total, 190 different virus-specific T-cell populations were FACsorted using pMHC-tetramers, followed by a short-term in vitro stimulation. The CDR3β nucleotide-sequences were determined using next-gen Illumina sequencing. CDR3β nucleotide-sequences that occurred at a frequency of less than 0.1% in each sample were excluded. CDR3β nucleotide-sequences that were identical and present in two different specificities, but present at high frequencies in one specificity, were only removed from the specificity that contained the sequences at very low frequencies (0.1-0.5%; n=135). CDR3β nucleotide-sequences that were identical and present in two different specificities at low frequency were considered contamination and removed from the library (82 sequences, 41 different-sequences). The numbers of different CDR3β AA-sequences that were encoded by the CDR3β nucleotide-sequence are shown at protein level. We then assessed how many CDR3β-AA-sequences were found in multiple individuals (shared) and how many were only found in a single individual (unique).