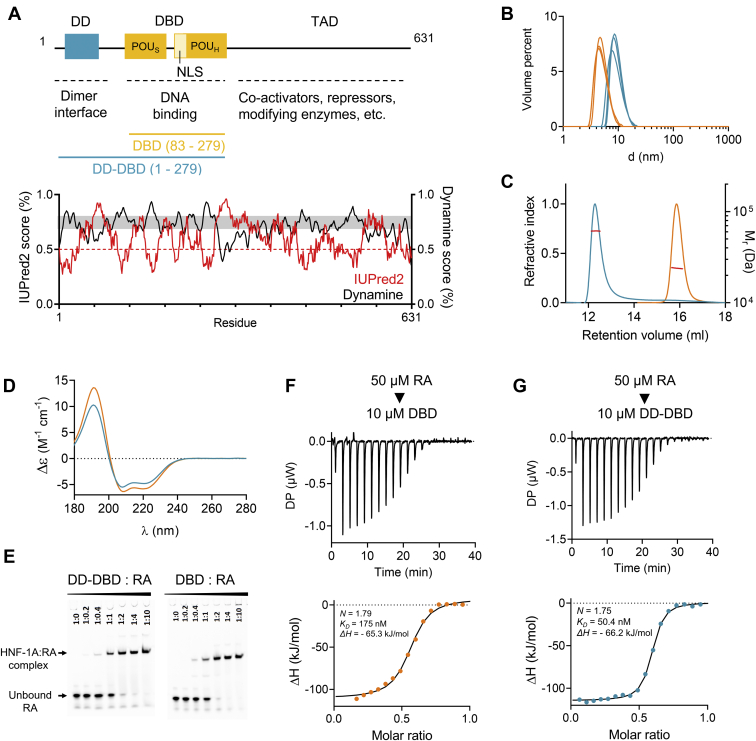
Figure 1.
Biophysical and functional characterization of HNF-1A DBD and DD-DBD constructs. A, top: domain overview of HNF-1A with residue numbers and indicated domain functions. DD-DBD (residues 1–279) and DBD (residues 83–279) constructs used for in vitro protein characterization are shown in turquoise and orange, respectively. Bottom: IUPred2 and DynaMine prediction scores for full-length HNF-1A. IUPred2 scores above 0.5 indicate propensity for disorder, while scores below 0.5 predict ordered regions (54). DynaMine scores below 0.7 indicate disorder, while scores above 0.8 predict order. DynaMine prediction in the gray zone (scores between 0.7 and 0.8) indicate residues with highly context-dependent dynamics (55). B, DLS of DBD and DD-DBD. C, SEC-MALS profiles for DBD and DD-DBD. Red line: molecular weight based on MALS and refractive index. D, SRCD spectra for DBD and DD-DBD. E, EMSA titration native gels showing the complex formation of RA and respective HNF-1A construct. F and G, ITC titration curves for DBD and DD-DBD, respectively. Titrant: 50 μM double-stranded RA oligonucleotide. Titrand: 10 μM DBD (F) or DD-DBD (G). DBD, DNA-binding domain; DD, dimerization domain; DLS, dynamic light scattering; HNF-1A, Hepatocyte nuclear factor 1A; SEC-MALS, size-exclusion chromatography–multi-angle light scattering; SRCD, synchrotron radiation circular dichroism.