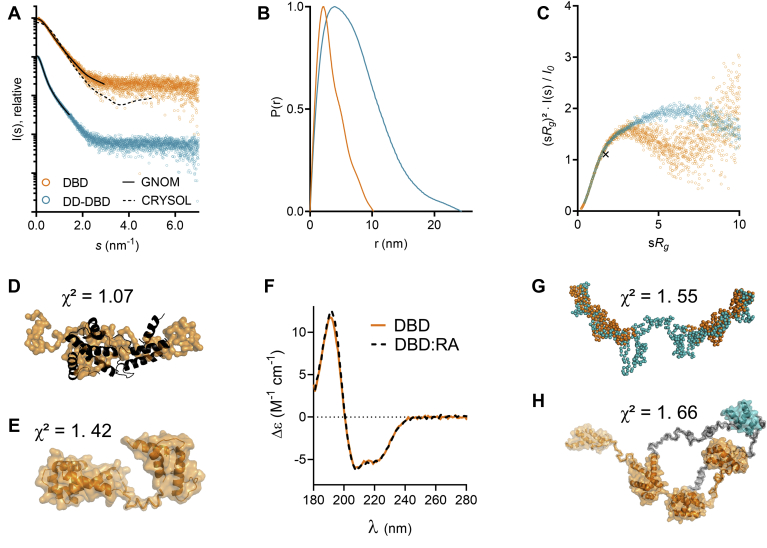
Figure 2.
SAXS data for DBD and DD-DBD.A, scattering curves for DBD and DD-DBD. Solid line: GNOM fit. Dashed line: CRYSOL fit with crystal structure model from PDB 1IC8 (16) (χ2 = 10.4). B, distance distribution functions for DBD and DD-DBD. C, dimensionless Kratky plot for DBD and DD-DBD, with the cross indicating the expected maximum for a globular particle (√3, 1.104) (61). D, GASBOR model of DBD (orange), superposed with the oligo-bound model from DBD:RA crystal structure (16). E, CORAL model for DBD. F, SRCD spectra for DBD in the presence and absence of RA. G, GASBOR model for DD-DBD (turquoise), overlaid with GASBOR model of DBD (orange) at estimated positions. H, CORAL model of DD-DBD. DBD residues are shown in orange, linker residues in gray, and DD residues in turquoise. DBD, DNA-binding domain; DD, dimerization domain; RA, rat albumin promoter oligonucleotide; SAXS, small-angle X-ray scattering; SRCD, synchrotron radiation circular dichroism.