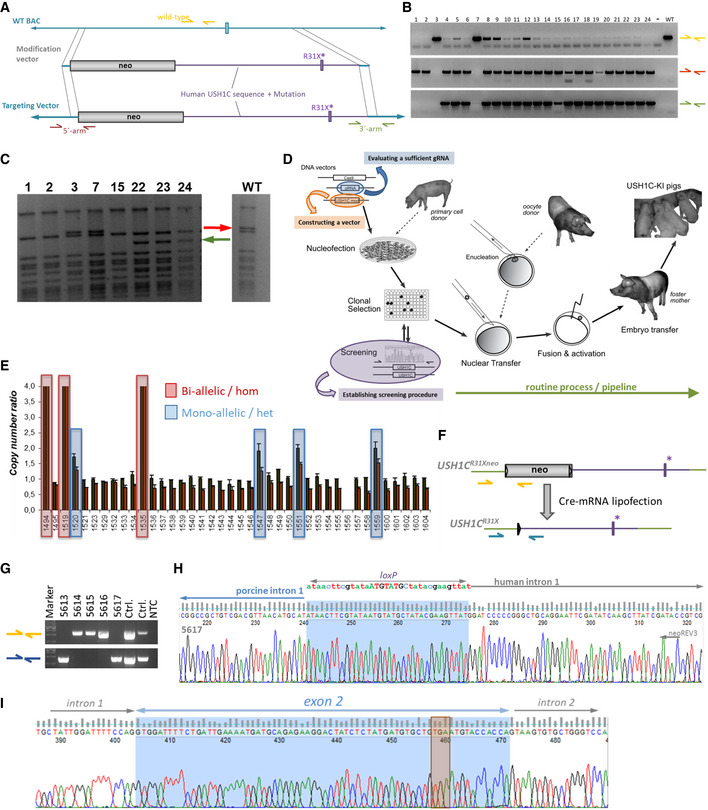
BAC clone CH242‐515C3 was modified by bacterial recombineering with a modification vector comprising the intended 1.5 kb human USHR31X segment (magenta) and a floxed neo selection cassette. Arrows represent primer for endpoint PCR to screen for correct recombination.
Correctly modified BAC clones are characterized by negative wild‐type PCR (yellow) and positive PCR spanning across the 5′‐arm (brown) and 3′‐arm (green) of homology.
XbaI digest of BAC clones for confirming integrity of the clone and verified the disappearance of an 18,779bp band (red arrow) and the appearance of a 14,194 bp band (green arrow) and a 5,020 bp band (too faint for detection).
Modified BAC vectors were co‐transfected with plasmids expressing Cas9 and gRNA4 into pig cells. Clonal selection for neomycin‐resistance delivered single cell clones (SSCs) which were expanded for screening and subsequent usage in SCNT.
SSCs were screened for modification by a qPCR‐based loss‐of‐wild‐type‐allele approach. Inverse copy number ratios of the USH1C locus vs reference sites in the POU5F1 and NANOG genes indicated SSCs with bi‐allelic (red boxes) and mono‐allelic (blue boxes) modification. Each qPCR was run in a duplicate, facilitating the calculation of 4 quotients of reference site : target site copy numbers for each sample. The mean value of these quotients are given ± SD. For ensuring characterization of SSCs, 2 independent reference sites were used and SSCs that appeared modified in the first screening were used in an independent experiment for confirmation.
Before SCNT, SSCs were lipofected with Cre‐encoding mRNA to remove the neo selection cassette. Primer sets were designed for discriminating founder animals with (yellow arrows) and without (blue arrows) neo.
Representative genotyping of a founder litter, indicating that animals 5613 and 5617 have sufficiently excised neo. For controls (Ctrl.), WT genomic DNA was mixed with modified BAC for the neo‐PCR (yellow) and mixed with a modified BAC that had been treated with Cre for the delta‐neo PCR (blue).
Representative electropherogram of PCR sequencing, confirming correct excision of neo in 5617. As a consequence of Cre‐mediated excision, a single lox‐site remains at the junction between porcine and human sequence.
Sanger sequencing confirmed also abundance of a correctly modified humanized exon 2. The orange box indicates the TGA‐nonsense codon causing the R31X mutation.