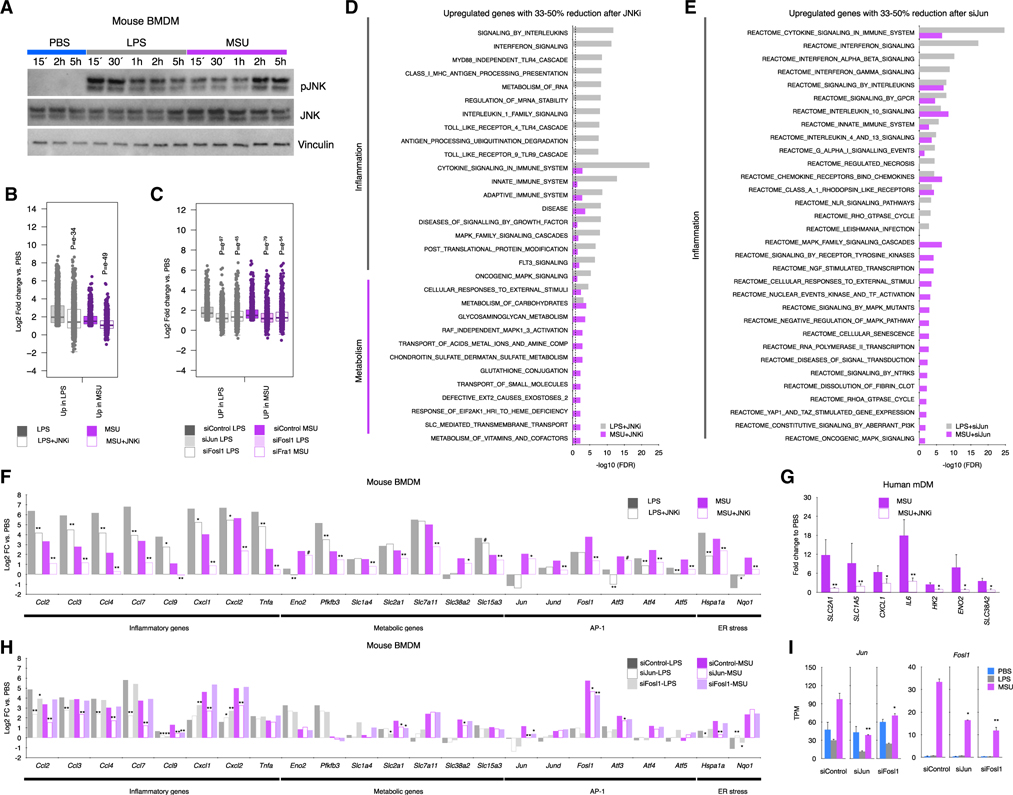
Figure 5. The inflammatory and metabolic program induced by MSUc is regulated through JNK.
(A) Protein analysis by WB of JNK and pJNK expression in BMDMs treated with LPS (100 ng/mL) or MSUc (250 μg/mL) at various time points. Data show prolonged pJNK expression in BMDMs treated with MSUc.
(B and C) Boxplots showing amelioration of gene expression after treatment with JNKi (20 μM) versus vehicle of genes upregulated by LPS or MSUc (B), genes upregulated only in LPS o rMSUc or commonly upregulated in LPS and MSUc for 5 h (C) (n ≥ 5/group). Data represent the distribution of fold change of upregulated genes in MSUc versus PBS or LPS versus PBS in cells treated with MSU + JNKi or LPS + JNKi.
(D) Gene ontology analysis using REACTOME of genes between 33% and 50% reduction of expression in MSUc + JNKi versus MSUc or LPS + JNKi versus LPS. Data show enrichment in inflammatory gene sets in LPS and MSUc and metabolic gene sets in MSUc.
(E) Gene ontology analysis using REACTOME of genes between 33% and 50% reduction of expression in siJun-MSUc versus siControl-MSUc or siJun-LPS versus siControl-LPS. Data show enrichment in inflammatory gene sets in LPS and MSUc gene sets.
(F–H) Expression analysis by RNA-seq (F and H) or qRT-PCR (G) of inflammatory and metabolic genes induced by MSUc or LPS showing reduction after treatment with JNKi in BMDMs (F) (n ≥ 5/group) or mDMs (G) (n = 3 donors/group), or in BMDMs after treatment with siJun or siFosl1 (H).
(I) Expression analysis by RNS-seq of Jun or Fosl1 in cells incubated with siControl or siJun or siFosl1 showing downregulation of Jun or Fosl1 in siJun or siFosl1 (#p < 0.10, *p < 0.05, **p < 0.01). Broken lines in (D and E) represents the cutoff for significance −log10(0.05) = 1.30.