Summary
MicroRNAs (miRNAs) play vital roles in plant development and defence responses against various stresses. Here, we show that blocking miR1871 improves rice resistance against Magnaporthe oryzae and enhances grain yield simultaneously. The transgenic lines overexpressing miR1871 (OX1871) exhibit compromised resistance, suppressed defence responses and reduced panicle number resulting in slightly decreased yield. In contrast, the transgenic lines blocking miR1871 (MIM1871) show improved resistance, enhanced defence responses and significantly increased panicle number leading to enhanced yield per plant. The RNA‐seq assay and defence response assays reveal that blocking miR1871 resulted in the enhancement of PAMP‐triggered immunity (PTI). Intriguingly, miR1871 suppresses the expression of LOC_Os06g22850, which encodes a microfibrillar‐associated protein (MFAP1) locating nearby the cell wall and positively regulating PTI responses. The mutants of MFAP1 resemble the phenotype of OX1871. Conversely, the transgenic lines overexpressing MFAP1 (OXMFAP1) or overexpressing both MFAP1 and miR1871 (OXMFAP1/OX1871) resemble the resistance of MIM1871. The time‐course experiment data reveal that the expression of miR1871 and MFAP1 in rice leaves, panicles and basal internode is dynamic during the whole growth period to manipulate the resistance and yield traits. Our results suggest that miR1871 regulates rice yield and immunity via MFAP1, and the miR8171‐MFAP1 module could be used in rice breeding to improve both immunity and yield.
Keywords: miR1871, MIM1871, blast disease resistance, yield traits, MFAP1, PTI
Introduction
Plants contain a two‐perception‐layer innate immune system to protect themselves from the invasion of pathogens. The first layer of immunity is the pathogen‐associated molecular pattern‐ (PAMP‐) triggered immunity (PTI), which is activated following the recognition of pathogen‐derived PAMPs by the receptors of plants. For example, the bacterium‐derived PAMP flg22 could be recognized by Arabidopsis receptor flagellin sensing 2 (FLS2) (Felix et al., 1999; Gomez‐Gomez and Boller, 2000) or by rice receptor OsFLS2 (Takai et al., 2008). The fungus‐derived PAMP chitin could be recognized by Arabidopsis receptor chitin elicitor receptor kinase (CERK1) (Miya et al., 2007) or by rice receptors such as chitin elicitor binding protein (OsCEBiP), OsCERK1, lysin motif‐containing proteins 4 (LYP4), and LYP6 (Liu et al., 2012; Shimizu et al., 2010). The second layer of immunity is effector‐triggered immunity (ETI) triggered by the recognition of pathogen‐derived avirulent effectors by the plant cognate resistance (R) genes (Jones and Dangl, 2006).
MiRNAs are short non‐coding RNAs that suppress gene expression by mediating DNA methylation, or mRNA cleavage, or translational inhibition via binding to the reverse complementary DNA/RNA sequences (Yu et al., 2017). Increasing evidence shows that miRNAs and their target genes participate in the regulation of plant PTI and ETI (Li et al., 2019b). miR393 is the first miRNA identified as an actor involving in both PTI and ETI against Pseudomonas syringe pv. tomato (Pst) DC3000. miR393 is up‐regulated with the treatment of flg22 and suppresses the expression of TIR1, AFB2 and AFB3, which encodes the auxin receptors (Navarro et al., 2006). Moreover, miR393 is up‐regulated during ETI triggered by the recognition of the avirulent effector AvrRpt2 by the cognate R receptor RPS2 (Zhang et al., 2011). Subsequently, several miRNAs are characterized as the regulators in ETI. In Arabidopsis, miR472 is involved in ETI by targeting and suppressing the R genes encoding nucleotide‐binding site leucine‐rich repeat (NLR) receptors, such as RPS5, which recognizes the effector AvrPphB to activate ETI against P. syringe (Boccara et al., 2014). In tomato, miR482 and miR2118 target the R genes encoding disease resistance proteins with a nucleotide‐binding site (NBS) and leucine‐rich repeat (LRR) motifs (Shivaprasad et al., 2012). In Medicago, miR2109 targets a large gene family containing conserved NB‐LRR domains (Zhai et al., 2011).
Rice is the most important crop supporting half of the world population. Rice blast disease is one of the most serious diseases threatening global rice production. This disease is caused by the fungal pathogen Magnaporthe oryzae (M. oryzae) and can occur during the whole rice growth period. Research over the recent years has revealed that miRNAs play key roles in rice resistance against M. oryzae (Li et al., 2019b). For example, several miRNAs have been identified as positive regulators in rice blast disease resistance, including miR159, miR160, miR162, miR166k/h, miR7695, miR398b and miR812w (Campo et al., 2013, 2021; Chen et al., 2021; Li et al., 2014, 2019a, 2020; Salvador‐Guirao et al., 2018). In contrast, some other miRNAs negatively regulate rice resistance to blast fungus, such as miR156, miR164a, miR167, miR168, miR169, miR319b and miR1873 (Li et al., 2017; Wang et al., 2018c, 2021; Zhang et al., 2018; Zhao et al., 2020) (Zhang et al., 2020; Zhou et al., 2020).
miRNAs not only regulate rice immunity but also participate in the regulation of rice development and yield. For example, miR172 regulates rice maturity and yield traits by controlling flowering time and floral organ development (Lee et al., 2014; Wang et al., 2015; Zhu et al., 2009). miR393 is involved in rice development by controlling tiller development and flowering (Xia et al., 2012). miR397 regulates grain size, grain number and grain yield(Zhang et al., 2013).
Moreover, some miRNAs and their target genes control both yield traits and immunity. In Brassica, miR1885 directly targets a defence‐related R gene BraTNL1 and a photosynthesis‐related gene BraCP24, to maintain a trade‐off between development and immunity (Cui et al., 2020). In rice, miR162 enhances the resistance against M. oryzae but compromises grain yield with reduced filled grain numbers per plant, whereas blocking miR162 enhanced grain yield but compromises the blast disease resistance (Li et al., 2020; Zhang et al., 2015). miR156 penalizes yield and suppresses blast disease resistance by suppressing Ideal Plant Architecture 1 (IPA1), whereas the expression of a miR156‐insensitive IPA1 enhances both rice yield and resistance significantly (Jiao et al., 2010a; Miura et al., 2010; Wang et al., 2018a; Zhang et al., 2020). Blocking miR168 enhances yield and rice blast disease resistance by manipulating the whole miRNA network via AGO1 (Wang et al., 2021). Blocking miR396 improves rice blast disease resistance and yield with increased panicle branching and grain size, as well as enhances the expression of target genes growth‐regulating factors (GRFs) (Chandran et al., 2019; Che et al., 2015; Duan et al., 2015). Consistently, overexpression of these GRFs enhances blast disease resistance and leads to a different change in yield traits (Chandran et al., 2019; Che et al., 2015; Duan et al., 2015). The transgenic lines overexpressing miR444 showed reduced tillers and decreased resistance, whereas overexpressing a target gene, OsMADS57, results in increased tillers and enhanced resistance (Guo et al., 2013; Xiao et al., 2017). Therefore, some miRNAs could be used in rice breeding to improve both yield and resistance or improve resistance without penalty of yield.
miR1871 is a rice‐specific 24‐nt miRNA and responsive to abiotic or biotic stresses. During drought stress, the accumulation of miR1871 is enhanced in the flag‐leaves of two tolerant cultivars, Nagina 22 and Vandana, whereas decreased in the two sensitive cultivars, Pusa Basmati 1 and IR64, suggesting that miR1871 acting as a positive regulator in rice drought responses (Balyan et al., 2017). Besides, the accumulation of miR1871 is fluctuated following the infection of M. oryzae (Li et al., 2014), indicating that miR1871 is involved in rice blast disease resistance. However, whether miR1871 regulates rice resistance against M. oryzae, and the roles of miR1871 in the regulation of rice yield traits, is still unknown.
In this study, we constructed the transgenic lines overexpressing MIR1871, the transgenic lines blocking miR1871, the mutants of a target gene of miR1871, mfap1, the transgenic lines overexpressing MFAP1 and the transgenic lines overexpressing both MFAP1 and MIR1871. We examined the resistance and yield traits of these lines. Our results showed that blocking miR1871 improved the expression of MFAP1, and enhanced rice blast disease resistance and yield traits. We carried out RNA‐seq to explore miR1871‐regulated signalling pathways and examined the miR1871‐MFAP1 module‐regulated PTI responses. Our results indicate that the miR1871‐MFAP1 module could be used as regulators in rice breeding to improve both resistance and yield simultaneously.
Results
Overexpression of miR1871 in rice compromises blast disease resistance
In a previous study, we demonstrated that miR1871 is differentially responsive to M. oryzae in a susceptible accession Lijiang xin Tuan Heigu (LTH) and a resistance accession Pyricularia‐Kanto51‐m‐Tsuyuake (IRBLkm‐Ts). LTH is a susceptible cultivar sensitive to over 1300 regional isolates of M. oryzae worldwide (Lin et al., 2001). IRBLkm‐Ts is a resistant cultivar carrying a resistance (R) locus Pi‐km (Tsumematsu et al., 2000). In this study, we further examined the expression pattern of miR1871 following the inoculation of a virulent M. oryzae isolate GZ8. GZ8 was a GFP‐tagged isolate that derived from a paddy yard in the north of China and virulent to Nipponbare (Li et al., 2019a). LTH showed increased miR1871 abundance following the inoculation of GZ8 in comparison with the mock samples at 12 and 48 hours post‐inoculation (hpi), whereas displayed decreased abundance at 24 hpi. In contrast, IRBLkm‐Ts exhibited indistinctive changes in miR1871 accumulation at all examined time points (Figure S1). These results indicated that miR1871 was involved in rice–M. oryzae interaction.
We then investigated the role of miR1871 in rice immunity. We constructed the transgenic lines overexpressing miR1871 (OX1871), which showed significantly higher amounts of miR1871 than the Nipponbare (NPB) control (Figure 1a). We examined the sensitivity of OX1871 to three M. oryzae isolates GZ8, CRB‐01 and TJ‐1. CRB‐01 is an isolate derived from the northeast of China without the known avirulence genes identified (Wang et al., 2018b). TJ‐1 is an isolate derived from the paddy yard in Tong‐Jiang County, Sichuan basin, the southwest of China. OX1871 lines were more sensitive to the three M. oryzae isolates with larger disease lesions and supported more fungal growth than the Nipponbare control following punch or spray inoculation (Figure 1b‐c, Figure S2a‐b). These results indicated that miR1871 facilitated the colonization of blast fungus in rice plants.
Figure 1.
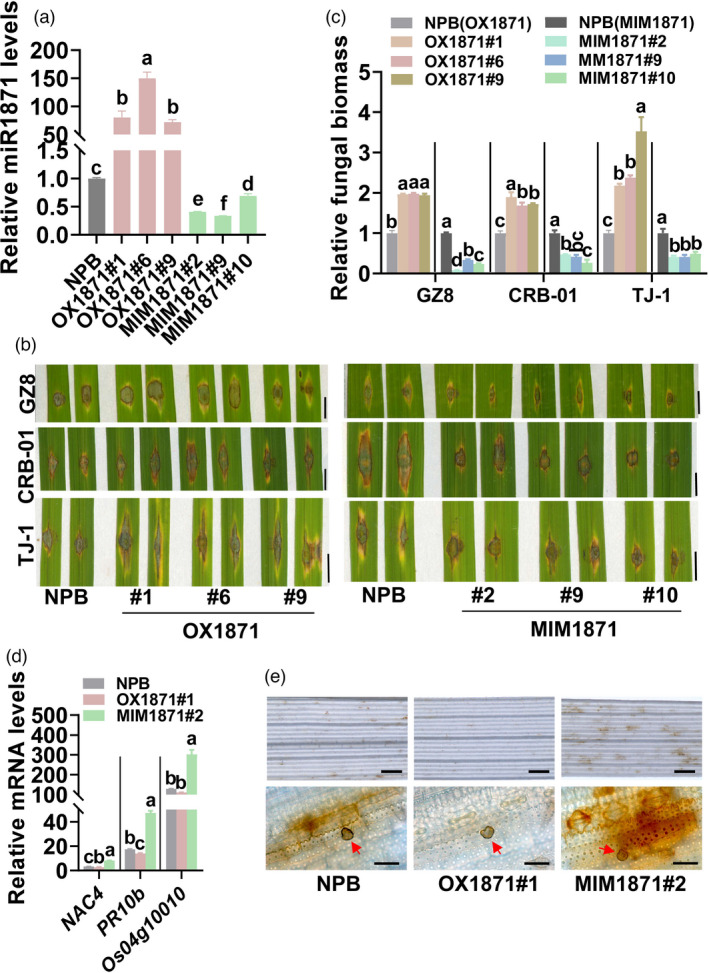
miR1871 regulates rice resistance against Magnaporthe oryzae. (a) The amounts of miR1871 in transgenic lines overexpressing the MIR1871 gene (OX1871), blocking miR1871 (MIM1871) and the Nipponbare (NPB) control. The levels of miR1871 were examined by quantitative reverse transcription polymerase chain reaction (qRT‐PCR). (b) Disease symptom on represented leaves of OX1871, MIM1871 and the Nipponbare control following inoculation of M. oryzae isolates GFP‐tagged Zhong8‐10‐14 (GZ8), CRB‐01 and TJ‐1, respectively. The photograph of OX1871 lines with their Nipponbare control was captured five days post‐inoculation (dpi). The photograph of MIM1871 lines with their Nipponbare control was captured seven days post‐inoculation (dpi). Size bar, 5 mm. (c) The relative fungal biomass of indicated isolates on OX1871, MIM1871 and the Nipponbare control. The fungal biomass was shown as the ratio of DNA levels of M. oryzae MoPot2 against DNA levels of rice ubiquitin. (d) The mRNA levels of defence‐related genes (NAC4, PR10b and Os04g10010) in OX1871, MIM1871 and the Nipponbare control at 12 hours post‐inoculation (hpi) of GZ8. The mRNA levels were normalized to that in Nipponbare. (e) The accumulation of hydrogen peroxide (H2O2) in OX1871, MIM1871 and the Nipponbare control at 48 hpi of GZ8. H2O2 was stained by 3,3’‐diaminobenzidine (DAB) and indicated by the intensity of brown. The photographs at the upper portion were taken with a stereomicroscope. Size bars, 1 mm. The photographs at the down portion were taken with a microscope (Zeiss imager A2). The red arrows indicate appressoria. Size bars, 40 µm. For a, c and d, data are shown as mean ± SD (n = 3 independent repeats), and different letters above the column indicate significant differences (P < 0.01) as determined by the one‐way Tukey–Kramer test. These experiments were repeated at least two times with similar results.
Blocking miR1871 enhances rice resistance against M. oryzae
As overexpressing miR1871 compromised rice blast disease resistance, we tried to explore whether blocking miR1871 could improve resistance. We then constructed the transgenic lines blocking miR1871 from binding its target genes by expressing a target mimic of miR1871 (MIM1871), which could bind miR1871 to form a double‐stranded complex (Figure S3a). MIM1871 lines displayed significantly decreased accumulation of miR1871 in comparison with the Nipponbare control (Figure 1a). As expected, MIM1871 lines showed enhanced resistance with smaller disease lesions and supported less fungal growth than the Nipponbare control after punch or spray inoculation (Figure 1b‐c, Figure S2a‐b). These results indicated that blocking miR1871 enhanced rice blast disease resistance.
As blocking miR1871 enhanced rice blast disease resistance, we examined whether miR1871 affected the defence responses triggered by blast fungus, including the induction of defence‐related genes and H2O2 accumulation. NAC domain‐containing protein 4 (NAC4), pathogenesis‐related genes 10b (PR10b) and Os04g10010 are defence‐response marker genes induced by M. oryzae (Li et al., 2020). MIM1871 displayed significantly enhanced expression of all these genes at 36 hpi of GZ8 in comparison with the Nipponbare control, whereas OX1871 exhibited decreased or unchanged expression of these genes (Figure 1d). Similarly, MIM1871 showed more accumulation of H2O2 in the invasive cells than that of the Nipponbare control at 48 hpi of GZ8, whereas OX1871 displayed less accumulation (Figure 1e). These results indicated that blocking miR1871 enhanced rice defence responses triggered by M. oryzae.
Blocking miR1871 enhances PTI‐related defence responses
To explore how blocking miR1871 enhanced rice blast resistance, we performed RNA‐seq experiments and compared the transcriptome profiles of MIM1871 and the Nipponbare control with three repeats to analyse the miR1871‐involved signalling pathways. The filtered data, including the numbers of total reads, mapped reads and the percentage of mapped reads, were listed in Table S1. 35,772 genes were identified (Table S2). Based on one‐way ANOVA analysis, 2,698 genes were significantly affected by miR1871 (P value < 0.05 and Log2 ˃1 or Log2<−1; Table S3, Figure S4). Gene Ontology (GO) analysis revealed that the up‐regulated genes in the MIM1871 line were enriched in pathways related to defence‐related hormone‐activated signalling, and responses to cold or salt stress, as well as responses to chitin treatment (Figure 2a, Table S4). To validate the RNA‐seq data, we selected ten genes that were clustered in more than two GO groups for expression analysis by quantitative reverse transcription polymerase chain reaction (qRT‐PCR). Consistently, all the ten genes were constitutively up‐regulated in MIM1871 (Figure 2b). Among these genes, five genes were clustered in the GO‐enriched genes that respond to chitin, including WRKY45, WRKY53, WRKY71, ERF020 and MAPKKK17. These results indicate that our RNA‐seq data are reliable, and blocking miR1871 constitutively up‐regulates rice defence‐related genes that may prime immune responses.
Figure 2.

Blocking miR1871 improves rice basal defence responses. (a) GO enrichment analysis of up‐regulated genes in MIM1871 in comparison with that in the Nipponbare control (NPB). The GO analysis was performed using the PANTHER (for protein annotation through evolutionary relationship) classification system (www.pantherdb.org) with default parameters. The 1918 up‐regulated genes in MIM1871 were used for analysis (Table S3). The classification results within the biological process categories were shown with P values from the statistical overrepresentation test. (b) Quantitative reverse transcription polymerase chain reaction (qRT‐PCR) data show the relative mRNA levels of the indicated genes in MIM1871 and the Nipponbare control. Data are shown as mean ± SD (n = 3 independent repeats). Different letters above the column show significant differences (P < 0.01) as determined by the one‐way Tukey–Kramer test. (c) The burst of reactive oxidative species (ROS) induced by flg22 in OX1871, MIM1871 and the Nipponbare control. Data are shown as mean ± SD (n = 6 independent repeats). (d) PAMPs (flg22 and chitin)‐induced callose deposition in the OX1871, MIM1871 and the Nipponbare control revealed by aniline blue staining. Size bar, 0.5 mm. (e) Quantitative analysis of PAMP‐induced callose deposition in (d). Data are shown as mean ± SD (n = 6 independent repeats). These experiments were repeated two times with similar results.
To explore the priming effect of blocking miR1871, we examined the PTI responses, such as the burst of reactive oxygen species (ROS) and callose deposition, in the transgenic lines and the Nipponbare control. The MIM1871 lines displayed higher induction of ROS in comparison with the Nipponbare control following the treatment of flg22, whereas the OX1871 lines showed the reversed phenotype (Figure 2c), suggesting a negative role of miR1871 in the regulation of flg22‐triggered ROS burst. As failed in detection of the chitin‐induced ROS in rice, we transiently expressed miR1871 or MIM1871 in Nicotiana benthamiana (N. benthamiana), respectively, to further examine the effects of miR1871 on ROS. With the treatment of flg22 and chitin, the leaves expressing miR1871 displayed suppressed ROS responses compared with the control leaves expressing GFP, whereas the leaves expressing MIM1871 showed enhanced ROS (Figure S5), indicating that blocking miR1871 enhanced PAMP‐triggered ROS. Moreover, both flg22 and chitin induced more callose deposits in the MIM1871 lines than that in the Nipponbare control, whereas induced fewer callose deposits in OX1871 lines (Figure 2d‐e). These results were consistent with the enrichment of genes that were responsive to chitin in the MIM1871 transgenic rice lines (Figure 2a), suggesting that blocking miR1871 could improve the PTI responses in rice.
miR1871 suppresses the expression of LOC_Os06g22850
As miRNAs regulate plant development and resistance by suppressing the expression of target genes, we then tried to explore the target genes of miR1871 to learn the signalling pathways controlled by miR1871. Three genes were predicted as the targets of miR1871, namely LOC_Os01g28300, LOC_Os06g22850 and LOC_Os11g19610 (http://plantgrn.noble.org/psRNATarget/; (Wu et al., 2009); Figure S3a). However, the mRNA levels of LOC_Os11g19610 were under the detecting threshold, and LOC_Os01g28300 was specifically expressed in seeds. LOC_Os06g22850 encodes an MFAP (hereafter named as MFAP1) and expressed in all rice organs (http://rice.plantbiology.msu.edu). We next explored the expression of MFAP1 in leaves of OX1871 and MIM1871. The mRNA levels of MFAP1 were significantly enhanced in all three examined MIM1871 lines, whereas only obviously suppressed in one of the three detected OX1871 lines (Figure S3b). miR1871 is a 24‐nt miRNA and is predicted to mediate DNA methylation via a DCL3‐dependent pathway (Wu et al., 2010). We then analysed the cytosine methylation pattern at the miR1871‐targeted site of MFAP1 in the Nipponbare control and a dcl3a mutant. We found that the methylation occurred in the Nipponbare control but not in the dcl3a mutant (Figure S3c), indicating that miR1871 controlled the cytosine methylation near the targeted sites. We then examined whether miR1871 suppressed the protein levels of MFAP1. The target site of MFAP1 was located at the intron of the 3’‐UTR region (Figure S3a). We made constructs expressing YFP‐fused 3’‐UTR of MFAP1, in which the target site of MFAP1 was located (35S: YFP‐UTRMFAP1 ) (Figure 3a). We co‐expressed the fused protein with miR1871 alone or with miR1871 and MIM1871 together. The YFP intensity and protein levels were decreased when the construct 35S: YFP‐UTRMFAP1 was co‐expressed with miR1871 in N. benthamiana, whereas were recovered when MIM1871 was added (Figure 3b‐c), indicating that miR1871 suppressed the expression of MFAP1, and MIM1871 could release the suppression by miR1871.
Figure 3.
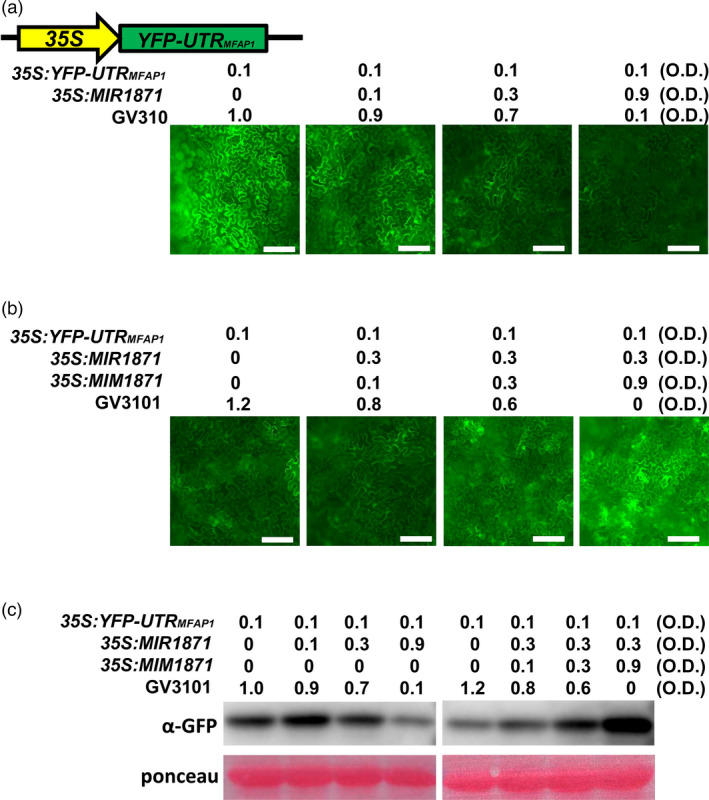
miR1871 suppresses the expression of MFAP1. (a) Confocal images of YFP‐UTRMFAP1 fluorescence signals suppressed by miR1871. The 35S: YFP‐UTRMFAP1 reporter construct was transiently expressed alone or co‐expressed with 35S: MIR1871 in Nicotiana benthamiana leaves by Agrobacterium‐mediated infiltration at indicated optical density (O.D.) concentrations. Size bars, 100 μm. (b) Confocal images of YFP‐UTRMFAP1 fluorescence signals co‐expressed with miR1871 and MIM1871. The 35S: YFP‐UTRMFAP1 reporter construct was transiently expressed alone or co‐expressed with 35S: MIR1871 and 35S: MIM1871 in N. benthamiana leaves by Agrobacterium‐mediated infiltration at indicated optical density (O.D.) concentrations. Size bars, 100 μm. GV3101 was used to complement the concentration of total bacteria. (c) Western blotting analysis showed the protein amounts of YFP in N. benthamiana leaves in (a). Protein extracted from the infiltrated leaves was subjected to Western blot analysis with anti‐GFP sera. The Ponceau‐stained Rubisco was used as the control indicating the equal sample loading. These experiments were repeated two times with similar results.
Mutations of MFAP1 compromise rice blast disease resistance and PTI responses
We next examined the role of MFAP1 in rice immunity. We constructed mutant lines of MFAP1 using the CRISPR/Cas9 technology in the Nipponbare background. We designed two gRNA sites for this gene to make sure that at least one gRNA is working (Figure S6a) and identified two homozygous mutants (mfap1 and mfap2). mfap1‐1 carried a 21‐bp deletion nearby gRNA1 site and a 1‐bp deletion nearby gRNA2 sites resulting in deletion of 7 amino acids (aa), and changes of several amino acids, as well as a premature stop (Figure S6b and d). mfap1‐2 carried an insertion of C nearby gRNA1 site and a 1‐bp deletion nearby gRNA2 sites resulting in changes from aa 57 and led to a premature stop after aa 70 (Figure S6c and e).
We then assessed the sensitivity of mfap1 mutants to M. oryzae. Consistent with the phenotype of OX1871, mfap1 mutants displayed compromised resistance against M. oryzae accompanied by bigger disease lesions and more fungal biomass than the Nipponbare control (Figure 4a‐b). Moreover, like OX1871, mfap1 mutants showed lower induction of ROS and fewer callose deposits than the Nipponbare control (Figure 4c‐f). These results indicated that miR1871 possibly repressed rice blast disease resistance by suppressing the expression of MFAP1, the mutations of which compromised rice immunity and PTI responses.
Figure 4.

Mutations of MFAP1 compromise rice resistance against Magnaporthe oryzae. (a) Blast disease lesions on the represent leaves of mfap1 and the Nipponbare (NPB) control at 5 days post‐inoculation with CRB‐01, GZ8 and RB22, respectively. Size bar, 5 mm. (b) The relative fungal biomass of indicated isolates in (a). The ratio of DNA levels of M. oryzae MoPot2 against DNA levels of rice ubiquitin was shown as the relative fungal biomass. Data are shown as mean ± SD (n = 3 independent repeats). (c) The accumulation of hydrogen peroxide (H2O2) in mfap1 and the Nipponbare control at 48 hours post‐inoculation (hpi) of M. oryzae isolates GZ8. H2O2 was stained by 3,3’‐diaminobenzidine (DAB) and indicated by the intensity of brown. The red arrows indicate appressoria. The photographs at the upper portion were taken with a stereomicroscope. Scale bars, 1 mm. The photographs at the down portion were taken with a microscope (Zeiss imager A2). Scale bars, 40 µm. (d) The burst of reactive oxidative species (ROS) induced by flg22 in mfap1 and the Nipponbare control. Data are shown as mean ± SD (n = 6 independent repeats). (e) PAMPs (flg22 and chitin)‐induced callose deposition in mfap1 and the Nipponbare control revealed by aniline blue staining. Size bar, 0.5 mm. (f) Quantitative analysis of PAMP‐induced callose deposition in (e). Data are shown as mean ± SD (n = 6 independent repeats). For b and f, different letters above the column show significant differences (P < 0.01) as determined by the one‐way Tukey–Kramer test. These experiments were repeated two times with similar results.
MFAP1 contributes to MIM1871‐conferred blast disease resistance
To further study the roles of MFAP1 in miR1871‐regulated resistance against M. oryzae, we constructed the transgenic lines overexpressing MFAP1 in Nipponbare background (OXMFAP1) and in OX1871 background (OXMFAP1/OX1871), respectively. All the transgenic lines exhibited significantly increased expression of MFAP1 in comparison with the controls and the null segregants (Figure S7a‐c). Like MIM1871, the OXMFAP1 lines were more resistant than the Nipponbare control and the null segregants, associated with smaller lesions and lower fungal biomass (by ~50%) (Figure 5a‐b). Moreover, the OXMFAP1/OX1871 lines also showed enhanced resistance in comparison with the Nipponbare control, the OX1871 lines, and the null segregants (Figure 5a‐b). These results suggest that MFAP1 contributes to MIM1871‐conferred resistance to M. oryzae.
Figure 5.
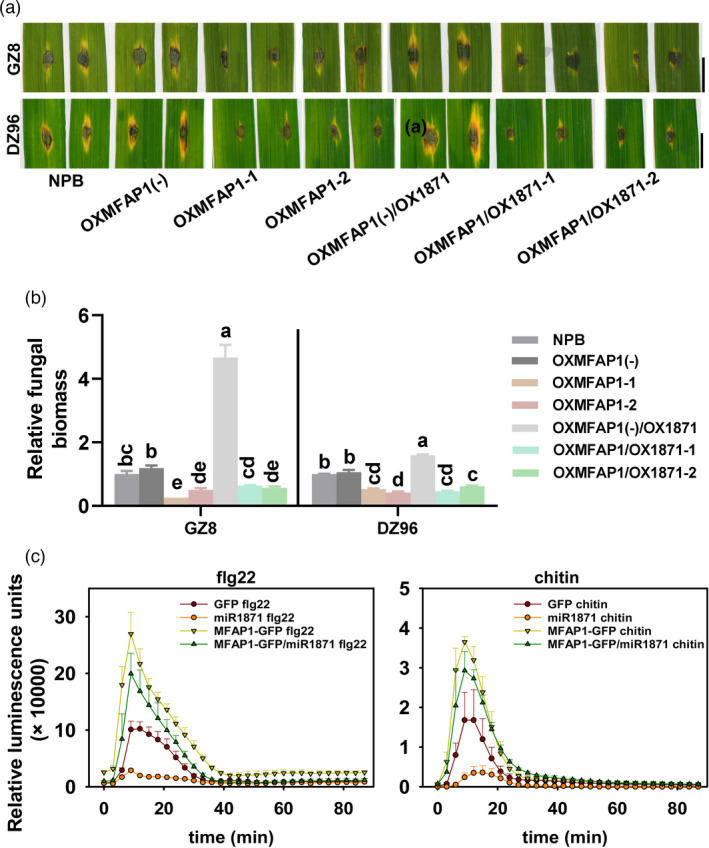
MFAP1 contributes to MIM1871‐regulated resistance and positively regulates basal defences. (a) The disease symptom on represented leaves of OXMFAP1, OXMFAP1/OX1871, the null segregants OXMFAP1(‐) and OXMFAP1(‐)/OX1871, and the Nipponbare control following the inoculation of M. oryzae isolates GFP‐tagged Zhong8‐10‐14 (GZ8) and DZ96, respectively. Size bar, 5 mm. The photograph was captured five days post‐inoculation (dpi). (b) The relative fungal biomass of indicated isolates in (a). The fungal biomass was shown as the ratio of DNA levels of M. oryzae MoPot2 against DNA levels of rice ubiquitin. Different letters above the column show significant differences (P < 0.01) as determined by the one‐way Tukey–Kramer test. (c) The reactive oxidative species (ROS) burst induced by flg22 and chitin in the leaves of N. benthamiana transiently expressing YFP‐tagged MFAP1, YFP‐tagged MFAP1 and miR1871, miR1871 alone and YFP alone, respectively. Data are shown as mean ± SD (n = 6 independent repeats). These experiments were repeated two times with similar results.
We tried to explain how MFAP1 regulated rice immunity. We first tested the location of MFAP1 in plant cells by transient expression of a YFP‐fused MFAP1 (35S: MFAP1‐YFP) in N. benthamiana. The YFP signals were enriched nearby the propidium iodide (PI)‐dyed cell wall (Figure S8a). Plasmolysis assay further revealed that MFAP1 was in apoplast nearby the cell wall (Figure S8b).
As MFAP1 was located nearby the cell wall, and mfap1 mutants displayed compromised PTI responses, we anticipated that MFAP1 could enhance PTI responses. We transiently expressed GFP‐fused MFAP1 (MFAP1‐GFP) in N. benthamiana to examine the effect of MFAP1 on PTI responses. As anticipated, overexpression of MFAP1‐YFP led to higher ROS and more callose deposits triggered by flg22 and chitin, compared with the control that transient expression of YFP only (Figure S9a‐d). These results were consistent with the defence responses triggered by PAMPs in MIM1871. Moreover, when miR1871 and MFAP1‐YFP were co‐expressed, MFAP1‐YFP partially recovered miR1871‐suppressed ROS burst (Figure 5c), indicating that MFAP1 played a positive role in MIM1871‐conferred blast disease resistance by improving the PTI responses.
The miR1871‐MFAP1 module regulates rice agronomic traits and yield
Intriguingly, we found that the miR1871‐MFAP1 module also regulates rice yield. The yield of rice is determined by several components, including panicle number, grain number per panicle, seed setting rate (SSR) and grain weight. We then examined these parameters and some other yield traits of OX1871, MIM1871 and mfap1 planted in a paddy field in the rice‐growing season of 2019 and 2020. All the examined lines displayed similar plant architecture but shorter plants in comparison with the Nipponbare control (Figure 6a, Table S5). OX1871 and mfap1 generated fewer panicles, fewer grains per panicle, lower SSR but heavier grain weight resulting in a significant decrease in grain yield per plant (Figure 6b‐f, Table S5). In contrast, MIM1871 showed lower SSR but more panicles, more grains per panicle and heavier grains leading to a remarkable increase in yield per plant (Figure 6b‐f, Table S5). We then examined the expression of several genes characterized as the regulators of rice tiller numbers in OX1871 and MIM1871, namely OsLRK2, OsAAP5 and OsSPL14. While OsLRK2 acted as a positive regulator, OsAAP5 and OsSPL14 acted as negative regulators in tillering (Jiao et al., 2010b; Kang et al., 2017; Wang et al., 2019). Consistently, MIM1871 showed enhanced expression of OsLRK2 but suppressed expression of OsAAP5, whereas OX1871 and mfap1 mutant displayed a reverse expression pattern of the two genes (Figure 6g), indicating the involvement of the two genes in the miR1871‐MFAP1 module‐regulated tillering. OsSPL14 is a well‐known negative regulator of tiller number in rice (Jiao et al., 2010a; Miura et al., 2010; Wang et al., 2018a; Zhang et al., 2020). The expression of OsSPL14 was not significantly changed in all the examined lines in comparison with the Nipponbare control (Figure 6g), indicating OsSPL14 was not involved in miR1871‐mediated regulation of tiller number. Altogether, these results suggested that miR1871 possibly regulates these yield traits via MFAP1, and blocking miR1871 is helpful for the improvement of grain yield by enhancing panicle number and size.
Figure 6.

miR1871‐MFAP1 module regulates rice yield. (a) The gross morphology of OX1871, MIM1871, mfap1 and the Nipponbare (NPB) controls at the maturation stage. Scale bars, 50 cm. (b‐c) The panicle number, grain number per panicle and yield per plant of OX1871, MIM1871, mfap1 and the Nipponbare control planted in the rice‐growing season of 2020. Data are shown as mean ± SD (n = 5 independent plants). (e) Photograph of grains per plant of OX1871, MIM1871 and the Nipponbare control. Scale bars, 5 cm. (f) Photograph of grains per plant of mfap1 and the Nipponbare control. Scale bars, 5 cm. (g) The mRNA levels of tillering‐related genes in the basal internode of OX1871, MIM1871 and the Nipponbare control. Data are shown as mean ± SD (n = 3 independent samples). For b, c, d and g, different letters above the bars indicate a significant difference (P < 0.05 (b, c and d); P < 0.05 (g)) as determined by the one‐way Tukey–Kramer test.
miR1871‐MFAP1 module is dynamically expressed during the growing period
To find out how the “miR1871‐MFAP1” module controlling yield and immunity, we performed a time‐course experiment and examined the expression of miR1871 and MFAP1 in rice leaves, panicles and basal internode. The amounts of miR1871 were decreased gradually in leaves during the vegetative stage and in panicles during the productive stage (Figure 7a‐b). Moreover, miR1871 was decreased significantly in basal internode before tillering but increased remarkably after tillering (Figure 7c). Conversely, the mRNA levels of MFAP1 were increased accordingly during the vegetative and reproductive stages (Figure 7a‐b). This expression pattern of miR1871 and MFAP1 was consistent with the enhanced blast disease resistance and development of panicles in mature rice plants. When the rice plants grew up, the amounts of miR1871 were decreased gradually in leaves and panicles leading to enhanced expression of MFAP1, which improved blast disease resistance and panicle formation (Figure 7d). Moreover, during the tillering stage, the decreased miR1871 amounts in basal internode led to the increase in tiller number, whereas after tillering stage, the increased miR1871 suppressed the tillering to avoid the invalid tiller (Figure 7c‐d). Thus, the dynamical and spatial expression of miR1871 fine‐tunes rice immunity and yield via MFAP1. However, the mRNA levels of MFAP1 in the basal internode were not reversely consistent with the amounts of miR1871 before and after tillering (Figure 7c). Such an inconsistency indicated that MFAP1 is also regulated by other regulators, or there existed other target genes involved in the regulation of panicle number.
Figure 7.

miR1871 and MFAP1 dynamically and spatially expressed during the rice‐growing period. (a) The accumulation of miR1871 and the mRNA levels of MFAP1 in leaves of the Nipponbare plants during the vegetative stage. (b) The accumulation of miR1871 and the mRNA levels of MFAP1 in panicles of the Nipponbare plants during the reproductive stage. (c) The accumulation of miR1871 and the mRNA levels of MFAP1 in basal internode before and after tillering. For a‐c, data are shown as mean ± SD (n = 5 independent plants). Different letters above the bars indicate a significant difference (P < 0.01) as determined by the one‐way Tukey–Kramer test. (d) A model of miR1871 regulates the expression of MFAP1 throughout the rice growth period. During the whole growth period, miR1871 is gradually decreased and the levels of MFAP1 are increased to enhance disease resistance and improve panicle development.
Discussion
miRNAs fine‐tune the trade‐off between plant immunity and yield. In this work, we reported that blocking miR1871 improved both immunity and yield. While OX1871 exhibited compromised blast disease resistance and reduced grain yield (Figures 1 and 6), MIM1871 showed enhanced resistance and increased grain yield compared with the Nipponbare control (Figures 1 and 6). Moreover, consistent with the resistance phenotype of OX1871, mfap1 displayed the compromised resistance and grain yield (Figure 4). Conversely, OXMFAP1 and OXMFAP1/OX1871 lines all displayed enhanced resistance like MIM1871 (Figure 5). These results indicated that the miR1871‐MFAP1 module was an important regulator for rice resistance and yield traits and could be used in rice breeding to improve resistance and yield simultaneously.
Intriguingly, the miR1871‐MFAP1 module regulated PTI responses. Overexpression of miR1871 or mutations of MFAP1 resulted in suppressed induction of ROS and callose deposition. Conversely, blocking miR1871 or overexpression of MFAP1 enhanced these defence responses (Figures 2, 4 and 5). Moreover, blocking miR1871 constitutively enhanced the expression of genes responsive to chitin (Figure 2), which in turn primes PAMP‐triggered immune responses. These results indicated that the miR1871‐MFAP1 module controls rice immunity by regulating PTI responses. However, current data did not address whether the miR1871‐MFAP1 module participated in the regulation of ETI against M. oryzae, which could be an interesting future research focus.
MFAP1 protein was first identified as the extracellular matrix glycoproteins in animals (Horrigan et al., 1992) and later found to be involved in spliceosome preparations (Neubauer et al., 1998). MFAP proteins play important roles in microfibril assembly, tissue homeostasis, cell viability and tumour progression. MFAPs contain five subfamily members, namely from MFAP1 to MFAP5. MFAP1 was first cloned in the human genome and characterized as a regulator of inherited diseases affecting microfibrils (Yeh et al., 1994). In the fungus Cenangium ferruginosum, MFAP1 is identified as a growth‐related marker gene (Ryu et al., 2018). In Drosophila, a homolog of MFAP1 was identified as a nucleolar protein involving in pre‐messenger RNA processing, spliceosome assembly and cell survival (Rode et al., 2017; Ulrich et al., 2016). In C. elegans, a mfap1 gene encodes a nuclear‐located splicing factor that can affect alternative splicing of mRNAs (Ma et al., 2012). In this study, we demonstrated that the rice MFAP1 protein was located near the cell wall. This location resembles the location of some animal MFAP family members in animals, which are in the extracellular matrix (Horrigan et al., 1992). Moreover, the location of the MFAP1 protein was consistent with its roles in physical defence responses such as callose deposition (Figure S9), the component of which was β‐1,3 glucan and located on the cell wall. However, it is still unclear how MFAP1 participates in the regulation of ROS burst.
miR1871‐MFAP1 module also regulated yield traits (Figure 6). Both OX1871 and mfap1 mutants exhibited decreased grain yield due to decreased panicle number, grain number per panicle and SSR (Figure 6, Table S5). Conversely, MIM1871 displayed the reverse phenotype. These results indicated that miR1871 regulated rice yield traits via MFAP1. Further study about the agronomic traits of OXMFAP1 and MFAP1/OX1871 would greatly contribute to our understanding of the roles of the miR1871‐MFAP1 module. Moreover, how the cell wall‐located MFAP1 contributes to rice yield traits is unclear. Screening the interacting proteins of MFAP1 and learning about their biochemical function would be helpful to reveal how MFAP1 functioned in rice immunity and growth.
MFAP1 was possibly not the only target gene of miR1871. For example, Os01g28300 and Os06g19610 were also predicted as the targets of miR1871 (Wu et al., 2009). LOC_Os01g28300 encodes an F‐box domain‐containing protein, and LOC_Os11g19610 encodes a retrotransposon. However, both genes are not expressed in leaves or under the detecting threshold, indicating an impossibility of involvement of rice leave resistance. Further study is required to test whether these genes were targets of miR1871 and involved in the regulation of yield traits. For example, LOC_Os01g28300 is specifically expressed in seeds (http://rice.plantbiology.msu.edu). Whether LOC_Os01g28300 participates in miR1871‐regulated seed development needs further study.
Methods
Plant materials and growth conditions
We use LTH and IRBLkm‐Ts to examine the expression pattern of miR1871. We use rice (Oryza sativa) japonica accession Nipponbare (NPB) to construct the miR1871 overexpressing lines (OX1871), miR1871 blocking lines by expressing a target mimic of miR1871 (MIM1871), mfap1 mutants, MFAP1 overexpressing lines (OXMFAP1) and the double overexpressing lines (OXMFAP1/OX1871). For resistance and defence response assay, the transgenic lines and the Nipponbare control were planted in a glasshouse at 28 ± 2 with 70% relative humidity and 12‐h/12‐h light/dark cycles.
Trait measurements
For the analysis of agronomic traits, the transgenic lines and the controls were grown in a paddy yard in Wenjiang District, Sichuan Province, China, in the rice‐growing season from mid‐April to mid‐September in 2019 and 2020. The morphology of the rice lines was captured at the maturity stage. For each line, 30 rice plants were planted with a 15‐cm × 20‐cm of plant‐row spacing, and five or more individual plants were selected for yield trait analysis, including panicle number per plant, grain number per panicle, seed setting rate (the ratio of filled grains to the total grains per plant), 1,000‐grain weight and grain yield per plant. The 1,000‐grain weight and grain yield per plant were obtained after the filled grains were dried in a 42°C oven for one week using an SC‐A grain analysis system (Wanshen Ltd., Hangzhou, China). All the data were analysed by one‐way ANOVA followed by post hoc Tukey HSD analysis at P value < 0.05.
Plasmid construction and transformation
We amplified the genomic sequence of MIR1871 from the genomic DNA of Nipponbare with the specific primers miR1871‐F‐KpnⅠ and miR1871‐R‐SalⅠ containing 492 bp upstream and 457 bp downstream sequences of MIR1871 (Table S6). The plasmid 35S‐pCAMBIA1300 was used as the vector to express the MIR1871 gene promoted by constitutive promoter 35S (OX1871). We constructed a target mimic of miR1871 (MIM1871) by cloning a double‐stranded fragment reversely complementary to miR1871 with three inserted bases (AAACCAACATGATAACATCAGAGCCAT) into the KpnI‐SalI sites of the binary vector 35S‐pCAMBIA1300‐IPS1. The fragment substituted the target site of miR399 in the vector as described previously (Franco‐Zorrilla et al., 2007; Li et al., 2017) and acted as a target mimic to absorb miR1871 thus blocked the binding of miR1871 and its authentic target genes. The target mimic fragments were obtained by annealing with primers MIM1871‐F‐KpnI and MIM1871‐R‐SalI (Table S6). To construct the transgenic lines overexpressing MFAP1, we amplified the CDS of MFAP1 from cDNA of Nipponbare with the specific primers MFAP1‐F and MFAP1‐R. The fragment was inserted into the binary vector 35S‐pCAMBIA1300 to express the protein MFAP1. To make the transgenic lines overexpressing MFAP1 in OX1871, the CDS of the MFAP1 gene were cloned into the binary vector 35S‐pCAMBIA2300 (Table S6).
The CRISPR/Cas9 plasmid construct and mutant screen
We constructed the mutants of MFAP1 following previous reports (Li, W et al., 2017) with CRISPR (Clustered regularly interspaced short palindromic repeats)/Cas9 technique. The guide RNA sequences (Table S6) of MFAP1 were screened by a Cas‐OFFinder system (http://www.rgenome.net/cas‐offinder/) to prevent potential off‐target sites with screen parameters including less than three base‐pair mismatches and one DNA/RNA bulge. The maize ubiquitin promoter (UBI) upstream of a codon‐optimized hSpCas9 (Cong et al., 2013) was inserted into the binary vector pCAMBIA1300 with hygromycin selection (via hygromycin B phosphotransferase). The original BsaI site present in the pCAMBIA1300 backbone was removed using a point mutation kit (TransGen, China). A fragment containing an OsU6 promoter(Feng et al., 2013) and a negative selection marker gene ccdB flanked by two BsaI sites and an sgRNA derived from pX260(Cong et al., 2013) was inserted into this vector using an In‐fusion cloning kit (Takara, Japan) to produce the CRISPR/Cas9 binary vector pBGK032. E. coli strain DB3.1 was used for maintaining this binary vector. The 23 bp targeting sequences (including PAM) of MFAP1 were selected within the target genes, and their targeting specificity was confirmed using a Blast search against the rice genome (https://blast.ncbi.nlm.nih.gov/Blast.cgi; Hsu et al., 2013). The designed targeting sequences were synthesized and annealed to form the oligo adaptors. Vector pBGK032 was digested by BsaI and purified using a DNA purification kit (Tiangen, China). A ligation reaction (10 µL) containing 10 ng of the digested pBGK032 vector and 0.05 mM oligo adaptor was carried out and directly transformed to E. coli competent cells to produce CRISPR/Cas9 plasmids. Agrobacterium strain EHA105 was used to conduct rice genetic transformation. The genotype of transgenic lines was screened using hygromycin resistance analysis. To examine the mutation sites, we extracted the genomic DNA from the transgenic lines for PCR amplification using the primers flanking the designed target sites (Table S6). The PCR products (500‐600 bp) were sequenced and blasted to the WT genome sequence to identify the mutation sites.
Pathogen cultivation
We used Magnaporthe oryzae isolates, including enhanced GFP‐tagged Zhong8‐10‐14 (GZ8), CRB‐01, TJ‐1, DZ96 and RB22, to examine the resistance and defence responses of the transgenic lines and the Nipponbare control. The isolates were incubated on plates containing oatmeal and tomato agar media (OTA) at 28 °C with 12‐h/12‐h light/ dark cycles. After ten days, we scratched the hyphae growing on the medium and cultured the plates at 28 °C with constitutive 24‐hour light for sporulation. After five days, we collected the spores (1 × 105 spores/ mL) for resistance and defence response analysis.
Gene expression assay
We selected the rice samples from the indicated tissue at the indicated time points for gene expression assay by quantitative reverse transcription polymerase chain reaction (qRT‐PCR). For detection of the defence‐related gene expression, we extracted the total RNAs from the spray‐inoculated leaves of three‐ to five‐leaf‐stage plants at the indicated time points using TRIzol reagent (Invitrogen) and conducted the reverse transcription experiment using the PrimeScriptTM RT reagent Kit with gDNA Eraser (TaKaRa Biotechnology, China). We performed qRT‐PCR using SYBR Green mix (QuantiNova SYBR Green PCR Kit, Qiagen, China) and the indicated primers (Table S6). We selected the rice ubiquitin (UBQ) gene as the internal reference to normalize the expression of genes. For detection of miR1871 levels, we conducted the reverse transcription with total RNA and a miR1871‐specific stem‐loop RT primers (Table S6). We examined the amounts of miR1871 by qRT‐PCR with the specific primers (Table S6) and selected snRNA U6 as the internal reference to normalize the amounts of miR1871.
Inoculation and microscopy assay
We used the three‐ to five‐leaf‐stage seedlings for resistance and defence response assay. We inoculated the transgenic lines and the controls with or without the spores (1 × 105 spores/ mL) of indicated isolates by punch or spray inoculation. For resistance assays, we observed the symptom on the inoculated leaves at 5‐7 days post‐inoculation (dpi) and examined the fungal biomass as previously reported (Park et al., 2012). In brief, we extracted the DNA from the inoculated leaves and determined the relative fungal biomass by qRT‐PCR using the DNA levels of the M. oryzae MoPot2 gene against that of the rice Ubiquitin gene. For detection of H2O2 amounts, we collected the spray‐inoculated leaves at 36 hpi and placed the leaves in 3, 3’‐diaminobenzidine (DAB) (1 mg /ml, Sigma, Alorich, USA) for staining at room temperature without light treatment for 12 hours. Subsequently, we decoloured the stained leaves in 90% ethyl alcohol at 65°C several times (Xiao et al., 2003). The H2O2 accumulation was captured with a stereomicroscope (Zeiss imager A2).
Transient expression assay in Nicotiana benthamiana
We fused YFP with the full‐length 3’‐UTR of MFAP1 at its C‐terminus (35S: YFP‐ UTRMFAP1 ). The sequence of UTRMFAP1 was amplified from the Nipponbare genomic DNA (Table S6). We inserted the isolated fragment into KpnI‐SpeI sites of the binary vector 35S‐pCAMNIA1300‐YFP and got the binary vector 35S‐pCAMNIA1300‐YFP‐UTRMFAP1 . We transformed the vector into Agrobacterium strain GV3101 and conducted the agroinfection assay in N. benthamiana. In brief, we incubated the Agrobacterium strains harbouring the respective expression constructs (35: YFP‐UTRMFAP1 , 35S: miR1871, 35S: MIM1871) at 28°C overnight in LB media containing kanamycin (50 mg/ mL) and carbenicillin (50 mg/ mL) on a table shaking at 250 rpm. We collected the bacteria at 800 g for 5 min and resuspended them in an MMA buffer (10mM MES, 10mM MgCl2, 100 mM AS). We infiltrated the Agrobacteria into leaves of N. benthamiana for transient expression assay accordingly and examined the GFP intensity at 48 hpi by image acquisition using a NikonA1 Confocal Laser Scanning Microscope (Nikon Instruments, Inc., China). We performed Western blotting analyses to determine the amounts of YFP. In brief, we used 15 µg of total extracted protein to conduct the electrophoresis on a 10% SDS‐PAGE and transferred the proteins to a membrane for blot assay. The protein blot was reacted with 3000‐fold‐diluted anti‐GFP sera (Sangon Biotech, D110008, Shanghai, China), to detect YFP and actin accumulation.
PTI defence responses in rice and N. benthamiana
We examined the production of reactive oxygen species (ROS) with the leaves of three‐ to five‐leaf‐stage rice seedlings and leaves of N. benthamiana expressing miR1871, MIM1871, MFAP1, or MFAP1 and miR1871 together, respectively. Leaf strips were incubated in 200 µL water in a 96‐well plate for 12 hours and treated with 1 µM flg22 or 20 µg/ mL chitin in 200 µL buffer containing 20 mM luminol, 10 µg/ mL horseradish peroxidase (Sigma‐Aldrich Shanghai Trading Co Ltd, Shanghai, China). The production of ROS was determined as relative luminescence units using a GloMax96 Microplate Luminometer (Promega Biotech Co., Ltd, Beijing, China) for 30–60 min. To examine the PTI‐triggered callose deposition in N. benthamiana, the leaves expressing miR1871, or MIM1871, or MFAP1, or MFAP1 and miR1871 together were syringe‐infiltrated with 1 µM flg22 or 20 µg/ mL chitin for 16 hours. Then, the treated leaves were collected and stained with 0.01% aniline blue for half an hour following a previous report (Zhang et al., 2007). We examined callose deposition in rice following a previous report (Liu et al., 2012). The first leaves of the five‐day‐old seedling were treated with flg22 or chitin for 12 hours and then fixed in ethanol: acetic acid (3:1 [v/v]) solution for 5 h. The fixed leaves were then rehydrated successively in 70% and 50% ethanol for 2 h, respectively, and in water overnight. Then, the leaves were treated with 10% NaOH for 1 hour to make the tissues transparent. After washed three times with water, the leaves were incubated in the staining buffer containing 150 mM K2HPO4, pH 9.5, 0.01% aniline blue (Sigma‐Aldrich) for 4 hours. The images of callose deposition were captured with a fluorescence microscope under a UV channel (340 to 380 nm) (Zeiss imager A2.0), and the callose deposits were calculated using ImageJ software.
RNA‐seq and data analysis
We collected the leaf samples of MIM1871 and the Nipponbare control at the three‐leaf stage. We extracted the total RNAs of these samples and determined the RNA quantity and integrity by using NanoDrop 2000 (Thermo Scientific, Waltham, MA) and Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). RNA‐seq was conducted by Shanghai OE Biotech Co., Ltd. RNA‐seq libraries were constructed using the Illumina TruSeq RNA Library Prep Kit v2 (Illumina, San Diego, CA) following standard protocols and sequenced on an Illumina HiSeq X Ten platform, and 150 bp paired‐end reads were generated. Average 50 M read (Pair‐end 125bp model) for each library was output. The raw reads of fastq format were firstly processed using Trimmomatic (Bolger et al., 2014). All clean reads for each sample were aligned to a manufacture reference containing Oryza sativa. After reads mapping, a new GTF file was constructed by Cufflinks(Trapnell et al., 2012) and gene quantity was calculated by Htseq(Anders et al., 2015). Then, an R package DEseq2(Love et al., 2014) was applied to different gene expression analyses. Other data statistics and visualizing were performed by self‐R scripts. P value < 0.05 and fold change > 2 or fold change < 0.5 was set as the threshold for significantly differential expression. Hierarchical clustering analysis of DEGs was run with hclust in R version 3.2.0. GO enrichment and KEGG (Kanehisa et al., 2008) pathway enrichment analysis of DEGs were performed, respectively, using R based on the hypergeometric distribution.
Conflicts of interest
The authors declare no interest conflicts.
Author contributions
Y. L. and W‐M. Wang conceived the experiment, and together with T‐T. L., X‐R. H., Y. Z., Q. F., X‐M. Y. and X‐H. Z. carried it out; G‐B. L., J‐H. Zhao., J‐W. Zhang., Z‐X. Z., J. F. and Y. Huang analysed the data; S‐X. Z., Y‐P. J. and M. Pu carried out the field trial; Y. L. and W. Wang wrote the paper.
Supporting information
Figure S1 miR1871 is responsive to rice blast fungus Magnaporthe oryzae.
Figure S2 Blocking miR1871 enhances rice resistance against Magnaporthe oryzae following spray‐inoculation.
Figure S3 miR1871 is predicted to regulate the methylation of MFAP1 in rice.
Figure S4 The hot map of the genes differentially expressed in MIM1871 and the Nipponbare control detected by RNA‐seq.
Figure S5 miR1871 regulates PAMPs‐induced Burst of reactive oxidative species (ROS).
Figure S6 Mutation sites of MFAP1 in mfap1 mutants.
Figure S7 Overexpression of MFAP1 in the Nipponbare background and OX1871 background, respectively.
Figure S8 MFAP1 is located nearby the cell wall.
Figure S9 MFAP1 positively regulates PTI responses.
Table S1 The report of filtered data.
Table S2 The list of genes identified in MIM1871‐1 and the control NPB.
Table S3 The list of genes affected by miR1871.
Table S4 The GO profile of the up‐regulated genes in MIM1871 line
Table S5 The agronomic straits of OX1871, MIM1871, mfap1, and the NPB control in 2019 and 2020.
Table S6 The primers used in this study.
Acknowledgements
We thank Dr. Cai‐Lin Lei (Institute of Crop Science, Chinese Academy of Agricultural Sciences) for providing the monogenic resistant lines IRBLkm‐Ts. This work was supported by the National Natural Science Foundation of China (No. U19A2033, 31430072 and 32172417), the Department of Science and Technology of Sichuan Province (2020YJ0332 and 2021YJ0304) and the Open Research Fund of State Key Laboratory of Hybrid Rice (Hunan Hybrid Rice Research Center, 2021KF07).
Li, Y. , Li, T.‐T. , He, X.‐R. , Zhu, Y. , Feng, Q. , Yang, X.‐M. , Zhou, X.‐H. , Li, G.‐B. , Ji, Y.‐P. , Zhao, J.‐H. , Zhao, Z.‐X. , Pu, M. , Zhou, S.‐X. , Zhang, J.‐W. , Huang, Y.‐Y. , Fan, J. and Wang, W.‐M. (2022) Blocking Osa‐miR1871 enhances rice resistance against Magnaporthe oryzae and yield. Plant Biotechnol. J., 10.1111/pbi.13743
References
- Anders, S. , Pyl, P.T. and Huber, W. (2015) HTSeq–a Python framework to work with high‐throughput sequencing data. Bioinformatics, 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balyan, S. , Kumar, M. , Mutum, R.D. , Raghuvanshi, U. , Agarwal, P. , Mathur, S. and Raghuvanshi, S. (2017) Identification of miRNA‐mediated drought responsive multi‐tiered regulatory network in drought tolerant rice, Nagina 22. Sci Rep. 7, 15446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccara, M. , Sarazin, A. , Thiebeauld, O. , Jay, F. , Voinnet, O. , Navarro, L. and Colot, V. (2014) The Arabidopsis miR472‐RDR6 silencing pathway modulates PAMP‐ and effector‐triggered immunity through the post‐transcriptional control of disease resistance genes. PLoS Pathog, 10, e1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A.M. , Lohse, M. and Usadel, B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo, S. , Peris‐Peris, C. , Sire, C. , Moreno, A.B. , Donaire, L. , Zytnicki, M. , Notredame, C. et al. (2013) Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance‐associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 199, 212–217. [DOI] [PubMed] [Google Scholar]
- Campo, S. , Sanchez‐Sanuy, F. , Camargo‐Ramirez, R. , Gomez‐Ariza, J. , Baldrich, P. , Campos‐Soriano, L. , Soto‐Suarez, M. et al. (2021) A novel Transposable element‐derived microRNA participates in plant immunity to rice blast disease. Plant Biotechnol J, 19, 1798–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran, V. , Wang, H. , Gao, F. , Cao, X.L. , Chen, Y.P. , Li, G.B. , Zhu, Y. et al. (2019) miR396‐OsGRFs module balances growth and rice blast disease‐resistance. Front Plant Sci. 9, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che, R. , Tong, H. , Shi, B. , Liu, Y. , Fang, S. , Liu, D. , Xiao, Y. et al. (2015) Control of grain size and rice yield by GL2‐mediated brassinosteroid responses. Nat. Plants, 2, 15195. [DOI] [PubMed] [Google Scholar]
- Chen, J.F. , Zhao, Z.X. , Li, Y. , Li, T.T. , Zhu, Y. , Yang, X.M. , Zhou, S.X. et al. (2021) Fine‐tuning roles of Osa‐miR159a in rice immunity against Magnaporthe oryzae and development. Rice (N Y), 14, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong, L. , Ran, F.A. , Cox, D. , Lin, S. , Barretto, R. , Habib, N. , Hsu, P.D. et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science, 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, C. , Wang, J.J. , Zhao, J.H. , Fang, Y.Y. , He, X.F. , Guo, H.S. and Duan, C.G. (2020) A Brassica miRNA regulates plant growth and immunity through distinct modes of action. Mol. Plant. 13, 231–245. [DOI] [PubMed] [Google Scholar]
- Duan, P. , Ni, S. , Wang, J. , Zhang, B. , Xu, R. , Wang, Y. , Chen, H. et al. (2015) Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat Plants, 2, 15203. [DOI] [PubMed] [Google Scholar]
- Felix, G. , Duran, J.D. , Volko, S. and Boller, T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276. [DOI] [PubMed] [Google Scholar]
- Feng, Z. , Zhang, B. , Ding, W. , Liu, X. , Yang, D.L. , Wei, P. , Cao, F. et al. (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23, 1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco‐Zorrilla, J.M. , Valli, A. , Todesco, M. , Mateos, I. , Puga, M.I. , Rubio‐Somoza, I. , Leyva, A. et al. (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037. [DOI] [PubMed] [Google Scholar]
- Gomez‐Gomez, L. and Boller, T. (2000) FLS2: an LRR receptor‐like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell, 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Guo, S. , Xu, Y. , Liu, H. , Mao, Z. , Zhang, C. , Ma, Y. , Zhang, Q. et al. (2013) The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14 . Nat. Commun. 4, 1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrigan, S.K. , Rich, C.B. , Streeten, B.W. , Li, Z.Y. and Foster, J.A. (1992) Characterization of an associated microfibril protein through recombinant DNA techniques. J. Biol. Chem. 267, 10087–10095. [PubMed] [Google Scholar]
- Hsu, P.D. , Scott, D.A. , Weinstein, J.A. , Ran, F.A. , Konermann, S. , Agarwala, V. , Li, Y. et al. (2013) DNA targeting specificity of RNA‐guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, Y. , Wang, Y. , Xue, D. , Wang, J. , Yan, M. , Liu, G. , Dong, G. et al. (2010a) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. [DOI] [PubMed] [Google Scholar]
- Jiao, Y. , Wang, Y. , Xue, D. , Wang, J. , Yan, M. , Liu, G. , Dong, G. et al. (2010b) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kanehisa, M. , Araki, M. , Goto, S. , Hattori, M. , Hirakawa, M. , Itoh, M. , Katayama, T. et al. (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J. , Li, J. , Gao, S. , Tian, C. and Zha, X. (2017) Overexpression of the leucine‐rich receptor‐like kinase gene LRK2 increases drought tolerance and tiller number in rice. Plant Biotechnol. J. 15, 1175–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.S. , Lee, D.Y. , Cho, L.H. and An, G. (2014) Rice miR172 induces flowering by suppressing OsIDS1 and SNB, two AP2 genes that negatively regulate expression of Ehd1 and florigens. Rice (N Y), 7, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X.P. , Ma, X.C. , Wang, H. , Zhu, Y. , Liu, X.X. , Li, T.T. , Zheng, Y.P. et al. (2020) Osa‐miR162a fine‐tunes rice resistance to Magnaporthe oryzae and Yield. Rice (N Y), 13, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Cao, X.L. , Zhu, Y. , Yang, X.M. , Zhang, K.N. , Xiao, Z.Y. , Wang, H. et al. (2019a) Osa‐miR398b boosts H2O2 production and rice blast disease‐resistance via multiple superoxide dismutases. New Phytol. 222, 1507–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Jeyakumar, J.M.J. , Feng, Q. , Zhao, Z.X. , Fan, J. , Khaskheli, M.K. and Wang, W.M. (2019b) The roles of rice microRNAs in rice‐Magnaporthe oryzae interaction. Phytopathol. Res. 1, 33. [Google Scholar]
- Li, Y. , Lu, Y.G. , Shi, Y. , Wu, L. , Xu, Y.J. , Huang, F. , Guo, X.Y. et al. (2014) Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae . Plant Physiol. 164, 1077–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Zhao, S.L. , Li, J.L. , Hu, X.H. , Wang, H. , Cao, X.L. , Xu, Y.J. et al. (2017) Osa‐miR169 negatively regulates rice immunity against the blast fungus Magnaporthe oryzae . Front Plant Sci, 8, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Z.Z. , Jiang, W.W. , Wang, J.L. and Lei, C.L. (2001) Research and utilization of universally susceptible property of Japonica rice variety Lijiangxintuanheigu. Scientia Agricultura Sinjca, 34, 116–117. [Google Scholar]
- Liu, B. , Li, J.F. , Ao, Y. , Qu, J. , Li, Z. , Su, J. , Zhang, Y. et al. (2012) Lysin motif‐containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell, 24, 3406–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. and Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L. , Gao, X. , Luo, J. , Huang, L. , Teng, Y. and Horvitz, H.R. (2012) The Caenorhabditis elegans gene mfap‐1 encodes a nuclear protein that affects alternative splicing. PLoS Genet, 8, e1002827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, K. , Ikeda, M. , Matsubara, A. , Song, X.J. , Ito, M. , Asano, K. , Matsuoka, M. et al. (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–549. [DOI] [PubMed] [Google Scholar]
- Miya, A. , Albert, P. , Shinya, T. , Desaki, Y. , Ichimura, K. , Shirasu, K. , Narusaka, Y. et al. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad Sci. USA, 104, 19613–19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, L. , Dunoyer, P. , Jay, F. , Arnold, B. , Dharmasiri, N. , Estelle, M. , Voinnet, O. et al. (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science, 312, 436–439. [DOI] [PubMed] [Google Scholar]
- Neubauer, G. , King, A. , Rappsilber, J. , Calvio, C. , Watson, M. , Ajuh, P. , Sleeman, J. et al. (1998) Mass spectrometry and EST‐database searching allows characterization of the multi‐protein spliceosome complex. Nat. Genet. 20, 46–50. [DOI] [PubMed] [Google Scholar]
- Park, C.H. , Chen, S. , Shirsekar, G. , Zhou, B. , Khang, C.H. , Songkumarn, P. , Afzal, A.J. et al. (2012) The Magnaporthe oryzae effector AvrPiz‐t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen‐associated molecular pattern‐triggered immunity in rice. Plant Cell, 24, 4748–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rode, S. , Ohm, H. , Zipfel, J. and Rumpf, S. (2017) The spliceosome‐associated protein Mfap1 binds to VCP in Drosophila. PLoS One, 12, e0183733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, M. , Mishra, R.C. , Jeon, J. , Lee, S.K. and Bae, H. (2018) Drought‐induced susceptibility for Cenangium ferruginosum leads to progression of Cenangium‐dieback disease in Pinus koraiensis. Sci Rep, 8, 16368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador‐Guirao, R. , Hsing, Y.I. and San Segundo, B. (2018) The polycistronic miR166k‐166h positively regulates rice immunity via post‐transcriptional control of EIN2. Front Plant Sci, 9, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, T. , Nakano, T. , Takamizawa, D. , Desaki, Y. , Ishii‐Minami, N. , Nishizawa, Y. , Minami, E. et al. (2010) Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J, 64, 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad, P.V. , Chen, H.M. , Patel, K. , Bond, D.M. , Santos, B.A. and Baulcombe, D.C. (2012) A microRNA superfamily regulates nucleotide binding site‐leucine‐rich repeats and other mRNAs. Plant Cell, 24, 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai, R. , Isogai, A. , Takayama, S. and Che, F.S. (2008) Analysis of flagellin perception mediated by flg22 receptor OsFLS2 in rice. Mol. Plant Microbe Interact. 21, 1635–1642. [DOI] [PubMed] [Google Scholar]
- Trapnell, C. , Roberts, A. , Goff, L. , Pertea, G. , Kim, D. , Kelley, D.R. , Pimentel, H. et al. (2012) Differential gene and transcript expression analysis of RNA‐seq experiments with TopHat and Cufflinks. Nat Protoc. 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumematsu, H. , Yanoria, M.J.T. , Ebron, L.A. , Hayashi, N. , Ando, I. , Kato, H. , Imbe, T. et al. (2000) Development of monogenic lines of rice for blast resistance. Breed. Sci. 50, 229–234. [Google Scholar]
- Ulrich, A.K.C. , Seeger, M. , Schutze, T. , Bartlick, N. and Wahl, M.C. (2016) Scaffolding in the spliceosome via single alpha helices. Structure, 24, 1972–1983. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Li, Y. , Chern, M. , Zhu, Y. , Zhang, L.L. , Lu, J.H. , Li, X.P. et al. (2021) Suppression of rice miR168 improves yield, flowering time and immunity. Nat. Plants, 7, 129–136. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Wu, B. , Lu, K. , Wei, Q. , Qian, J. , Chen, Y. and Fang, Z. (2019) The amino acid permease 5 (OsAAP5) regulates tiller number and grain yield in rice. Plant Physiol. 180, 1031–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Zhou, L. , Shi, H. , Chern, M. , Yu, H. , Yi, H. , He, M. et al. (2018a) A single transcription factor promotes both yield and immunity in rice. Science, 361, 1026–1028. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Sun, S. , Jin, J. , Fu, D. , Yang, X. , Weng, X. , Xu, C. et al. (2015) Coordinated regulation of vegetative and reproductive branching in rice. Proc Natl Acad Sci U S A, 112, 15504–15509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W.W. , Liu, C.C. , Zhang, H.W. , Xu, H. , Zhou, S. , Fang, Y.L. , Peng, Y.L. et al. (2018b) Selection of differential isolates of Magnaporthe oryzae for postulation of blast resistance genes. Phytopathology, 108, 878–884. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y. , Xia, Y.Q. , Lin, S.Y. , Wang, Y.R. , Guo, B.H. , Song, X.N. , Ding, S.C. et al. (2018c) Osa‐miR164a targets OsNAC60 and negatively regulates rice immunity against the blast fungus Magnaporthe oryzae . Plant J. 95, 584–597. [DOI] [PubMed] [Google Scholar]
- Wu, L. , Zhang, Q.Q. , Zhou, H.Y. , Ni, F.R. , Wu, X.Y. and Qi, Y.J. (2009) Rice microRNA effector complexes and targets. Plant Cell, 21, 3421–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L. , Zhou, H. , Zhang, Q. , Zhang, J. , Ni, F. , Liu, C. and Qi, Y. (2010) DNA methylation mediated by a microRNA pathway. Mol. Cell, 38, 465–475. [DOI] [PubMed] [Google Scholar]
- Xia, K. , Wang, R. , Ou, X. , Fang, Z. , Tian, C. , Duan, J. , Wang, Y. et al. (2012) OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS One, 7, e30039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, S. , Brown, S. , Patrick, E. , Brearley, C. and Turner, J.G. (2003) Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid‐dependent amplification circuit is required for hypersensitive cell death. Plant Cell, 15, 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Z.Y. , Wang, Q.X. , Zhao, S.L. , Wang, H. , Li, J.L. , Fan, J. , Li, Y. et al. (2017) miR444b.2 regulates resistance to Magnaporthe oryzae and tillering in rice. Acta Phytppathol. Sin. 47, 511–522. [Google Scholar]
- Yeh, H. , Chow, M. , Abrams, W.R. , Fan, J. , Foster, J. , Mitchell, H. , Muenke, M. et al. (1994) Structure of the human gene encoding the associated microfibrillar protein (MFAP1) and localization to chromosome 15q15‐q21. Genomics, 23, 443–449. [DOI] [PubMed] [Google Scholar]
- Yu, Y. , Jia, T.R. and Chen, X.M. (2017) The 'how' and 'where' of plant microRNAs. New Phytol. 216, 1002–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, J. , Jeong, D.H. , De Paoli, E. , Park, S. , Rosen, B.D. , Li, Y. , Gonzalez, A.J. et al. (2011) MicroRNAs as master regulators of the plant NB‐LRR defense gene family via the production of phased, trans‐acting siRNAs. Genes Dev. 25, 2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D. , Liu, M. , Tang, M. , Dong, B. , Wu, D. , Zhang, Z. and Zhou, B. (2015) Repression of microRNA biogenesis by silencing of OsDCL1 activates the basal resistance to Magnaporthe oryzae in rice. Plant Sci. 237, 24–32. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Shao, F. , Li, Y. , Cui, H. , Chen, L. , Li, H. , Zou, Y. et al. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP‐induced immunity in plants. Cell Host Microbe. 1, 175–185. [DOI] [PubMed] [Google Scholar]
- Zhang, L.L. , Li, Y. , Zheng, Y.P. , Wang, H. , Yang, X. , Chen, J.F. , Zhou, S.X. et al. (2020) Expressing a target mimic of miR156fhl‐3p enhances rice blast disease resistance without yield penalty by improving SPL14 expression. Front Genet. 11, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Gao, S. , Zhou, X. , Chellappan, P. , Chen, Z. , Zhou, X. , Zhang, X. et al. (2011) Bacteria‐responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol. Biol. 75, 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Bao, Y. , Shan, D. , Wang, Z. , Song, X. , Wang, J. , He, L. et al. (2018) Magnaporthe oryzae induces the expression of a microRNA to suppress the immune response in rice. Plant Physiol. 177, 352–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y.C. , Yu, Y. , Wang, C.Y. , Li, Z.Y. , Liu, Q. , Xu, J. , Liao, J.Y. et al. (2013) Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 31, 848–852. [DOI] [PubMed] [Google Scholar]
- Zhao, Z.X. , Feng, Q. , Cao, X.L. , Zhu, Y. , Wang, H. , Chandran, V. , Fan, J. et al. (2020) Osa‐miR167d facilitates infection of Magnaporthe oryzae in rice. J. Integr. Plant Biol. 62, 702–715. [DOI] [PubMed] [Google Scholar]
- Zhou, S.X. , Zhu, Y. , Wang, L.F. , Zheng, Y.P. , Chen, J.F. , Li, T.T. , Yang, X.M. et al. (2020) Osa‐miR1873 fine‐tunes rice immunity against Magnaporthe oryzae and yield traits. J. Integr. Plant Biol. 62, 1213–1226. [DOI] [PubMed] [Google Scholar]
- Zhu, Q.H. , Upadhyaya, N.M. , Gubler, F. and Helliwell, C.A. (2009) Over‐expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biol. 9, 149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 miR1871 is responsive to rice blast fungus Magnaporthe oryzae.
Figure S2 Blocking miR1871 enhances rice resistance against Magnaporthe oryzae following spray‐inoculation.
Figure S3 miR1871 is predicted to regulate the methylation of MFAP1 in rice.
Figure S4 The hot map of the genes differentially expressed in MIM1871 and the Nipponbare control detected by RNA‐seq.
Figure S5 miR1871 regulates PAMPs‐induced Burst of reactive oxidative species (ROS).
Figure S6 Mutation sites of MFAP1 in mfap1 mutants.
Figure S7 Overexpression of MFAP1 in the Nipponbare background and OX1871 background, respectively.
Figure S8 MFAP1 is located nearby the cell wall.
Figure S9 MFAP1 positively regulates PTI responses.
Table S1 The report of filtered data.
Table S2 The list of genes identified in MIM1871‐1 and the control NPB.
Table S3 The list of genes affected by miR1871.
Table S4 The GO profile of the up‐regulated genes in MIM1871 line
Table S5 The agronomic straits of OX1871, MIM1871, mfap1, and the NPB control in 2019 and 2020.
Table S6 The primers used in this study.


