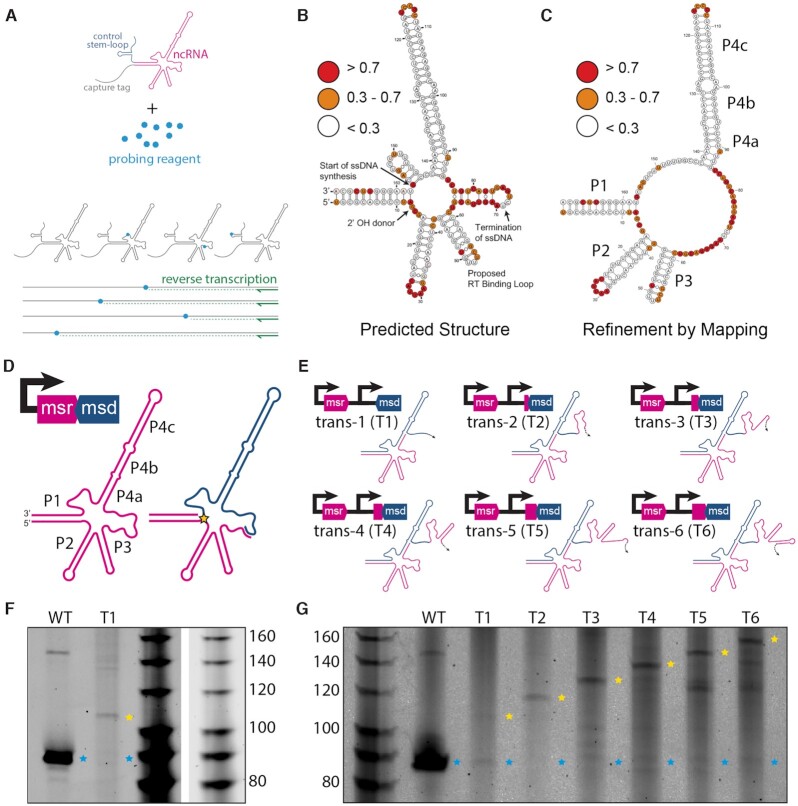
Figure 4.
Secondary structure in Eco1 ncRNA is not responsible for termination. (A) Schematic of chemical mapping experiment. (B) Chemical mapping binned reactivity on the in silico predicted secondary structure. Red indicates a reactivity value above 0.7, orange 0.3–0.7 and white below 0.3. The base that donates the -OH group to form the 2′-5′ linkage, the start of the template used for ssDNA synthesis, the canonical termination point and the proposed RT binding loop are indicated on the secondary structures with arrows. (C) Chemical mapping binned reactivity on a refined structure. (D) Eco1 ncRNA with verified stems labeled. Pink indicates the msr region and blue the msd region. The star indicates the 2′-5′ linkage. (E) Schematic of trans ncRNA architectures and operon architectures. Colors of schematics match ‘D’. (F) TBE-Urea gel of WT ncRNA and ncRNA trans-1 (T1). Blue stars show WT Retron-RNA size and yellow stars show an RT-DNA that does not terminate at the correct site. (G) TBE-urea gel of constructs T1-T6 using the same star nomenclature as 'F'.