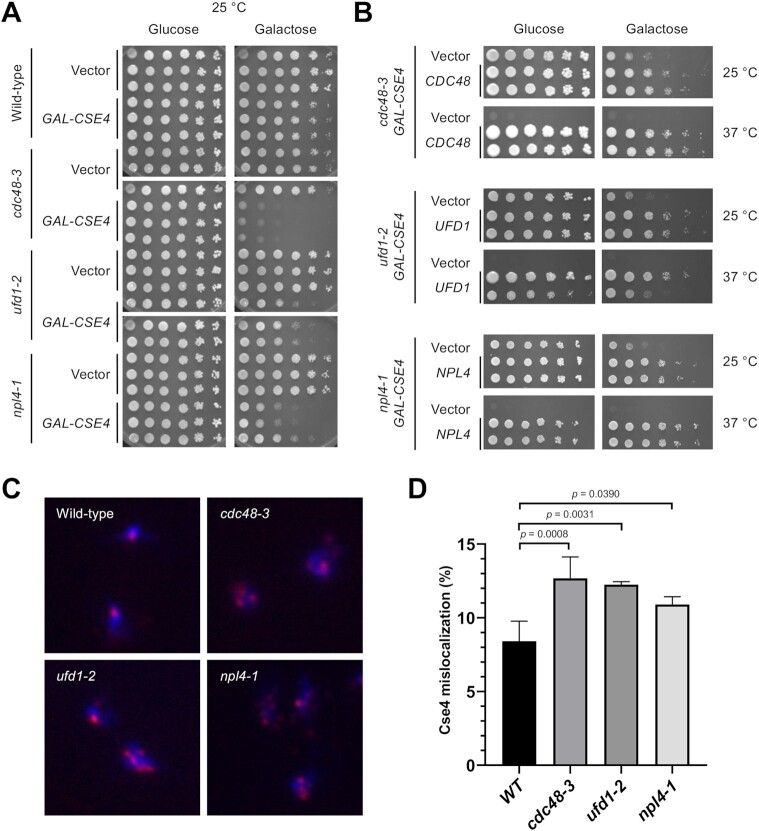
Figure 1.
Cdc48Ufd1/Npl4 segregase complex mutants exhibit SDL with GAL-CSE4 and show mislocalization of endogenous Cse4. (A) Overexpression of Cse4 results in SDL in cdc48-3, ufd1-2, and npl4-1 mutants. Wild-type, cdc48-3, ufd1-2 and npl4-1 cells containing either vector or GAL-CSE4 were spotted in five-fold serial dilutions on glucose (2%)- or galactose (2%)-containing synthetic medium selective for the plasmid. The plates were incubated at the permissive temperature of 25°C for 4 days. Three independent transformants for each strain are shown. (B) Plasmid (2μ)-borne CDC48, UFD1, and NPL4 can complement the GAL-CSE4 SDL phenotypes of cdc48-3, ufd1-2, and npl4-1 strains, respectively. Serial dilutions were conducted as described in A. The plates were incubated at 25°C and 37°C for 4–7 days. (C) Endogenous Cse4 is mislocalized in cdc48-3, ufd1-2 and npl4-1 strains. Localization of Cse4 was examined by chromosome spreads prepared from nocodazole (M) arrested wild-type, cdc48-3, ufd1-2 and npl4-1 cells. The cell cycle arrest was verified by FACS analysis (Supplementary Figure S3). DAPI (blue) and α-HA (red) staining were used to visualize DNA and Cse4 localization, respectively. Representative images of cells showing normal localization (counted as nuclei with one or two Cse4 foci) and mislocalization (counted as nuclei with more than two foci or a diffuse signal in the nucleus). (D) Quantification of Cse4 mislocalization as a percentage over total cell count. Error bars represent the standard deviation of multiple experiments. The increased mislocalization observed in the cdc48-3, ufd1-2 and npl4-1 strains was highly significant (one-way ANOVA, numbers of cells scored (n) = 627, 409, 418, 507, n = 503, 429, 340, 406, n = 349, 326, 555 and n = 330, 347, 448 for wild-type, cdc48-3, ufd1-2 and npl4-1, respectively in three or four independent experiments).