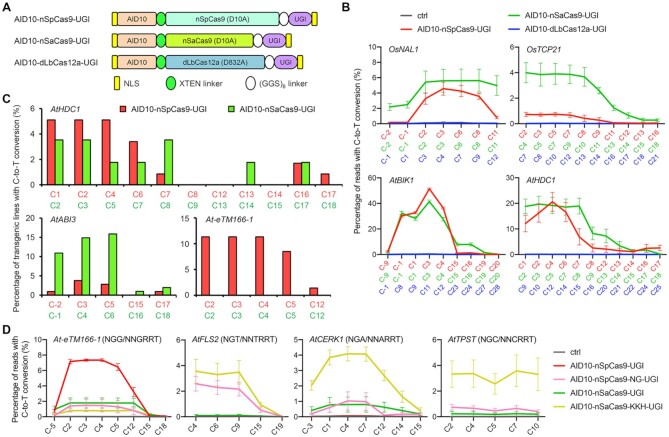
Figure 2.
Combination of AID10 with orthogonal or engineered Cas proteins expands the target scope. (A) Schematic diagram of AID10-based CBEs derived from SpCas9, SaCas9, and LbCas12a. (B) Comparison of cytosine editing efficiencies between different CBEs at endogenous genomic loci in rice (top) or Arabidopsis (bottom) protoplasts by amplicon deep sequencing. (C) Comparison of cytosine editing efficiencies between AID10-nSpCas9-UGI and AID10-nSaCas9-UGI at three endogenous genomic loci in transgenic T1 Arabidopsis plants. (D) Comparison of cytosine editing efficiencies between CBEs derived from PAM-relaxed Cas9 variants at endogenous genomic loci with canonical or non-canonical PAMs in Arabidopsis protoplasts by amplicon deep sequencing. The PAMs are shown in the parentheses. In (B) and (C), the numbers in red, green, and blue indicate protospacer positions for SpCas9, SaCas9, and LbCas12a, respectively. Data in (B) and (D) are shown as mean ± s.d. of two biological replicates performed at different times.