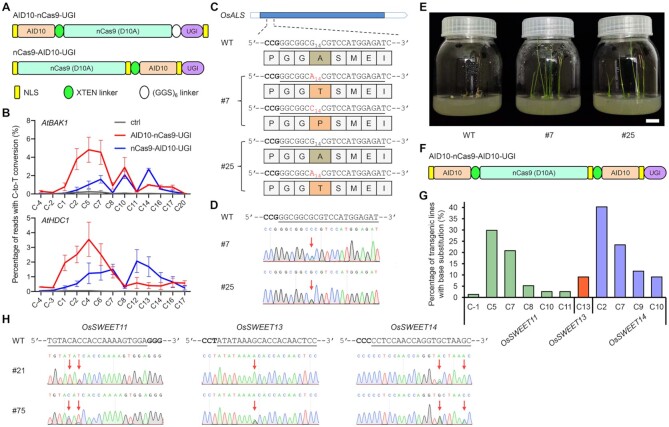
Figure 3.
nCas9-AID10-UGI and AID10-nCas9-AID10-UGI exhibit distinct activity windows relative to AID10-nCas9-UGI. (A) Schematic diagram of nCas9-AID10-UGI. (B) Comparison of cytosine editing efficiencies between nCas9-AID10-UGI and AID10-nCas9-UGI at endogenous genomic loci in Arabidopsis protoplasts by amplicon deep sequencing. Data are shown as mean ± s.d. of two biological replicates performed at different times. (C) Cytosine editing at the OsALS locus by nCas9-AID10-UGI in transgenic T0 rice lines #7 and #25 and resultant amino acid changes. The reverse complement of the target sequence is underlined and the PAM is in bold. The subscripted number indicates the protospacer position. (D) Sanger sequencing chromatograms show base conversions (marked by red arrows) described in (C). (E) Amino acid substitutions in OsALS confer herbicide resistance. The seedlings were germinated on the medium containing 0.1 mg/l imazethapyr for nine days. Scale bar = 2 cm. (F) Schematic diagram of AID10-nCas9-AID10-UGI. (G) Frequencies of cytosine editing at three endogenous genomic loci in transgenic T0 rice plants expressing AID10-nCas9-AID10-UGI and three sgRNAs, each targeting one indicated OsSWEET locus. (H) Sanger sequencing chromatograms show multiplex base conversions (marked by red arrows) by AID10-nCas9-AID10-UGI at the OsSWEET loci in two T0 rice lines.