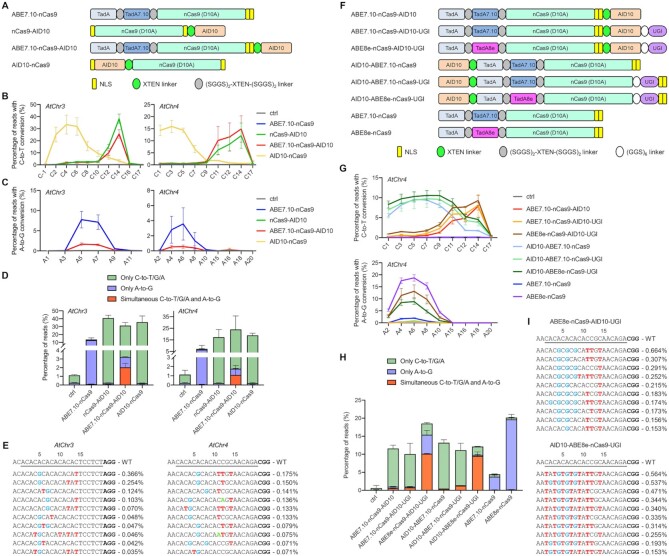
Figure 4.
Cytosine and adenine co-editing at the same target sequences by AID10-based dual base editors. (A) Schematic diagram of the dual base editor ABE7.10-nCas9-AID10. (B, C) ABE7.10-nCas9-AID10 mediates both C-to-T (B) and A-to-G (C) conversions at two endogenous genomic loci in Arabidopsis protoplasts as revealed by amplicon deep sequencing. Data are shown as mean ± s.d. of two biological replicates performed at different times. (D) Comparison of editing product compositions between ABE7.10-nCas9-AID10 and single-deaminase base editors. The percentage of indicated sequencing reads among total reads is presented as mean ± s.d. of two replicates. (E) The ten most abundant mutant alleles with concurrent cytosine and adenine substitutions by ABE7.10-nCas9-AID10 in one replicate of (D). Frequencies of indicated sequencing reads are shown on the right. The A-to-G, C-to-T, and C-to-A substitutions are in blue, red, and green, respectively. (F) Schematic diagram of eight dual base editors with different protein architectures. (G) Comparison of cytosine (top) and adenine (bottom) editing efficiencies between these base editors at the AtChr4 locus in Arabidopsis protoplasts by amplicon deep sequencing. Data are shown as mean ± s.d. of two biological replicates performed at different times. (H) Comparison of editing product compositions at the AtChr4 locus generated by these base editors. (I) The ten most abundant mutant alleles with concurrent cytosine and adenine substitutions by ABE8e-nCas9-AID10-UGI or AID10-ABE8e-nCas9-UGI in one replicate of (H). The A-to-G and C-to-T substitutions are in blue and red, respectively.