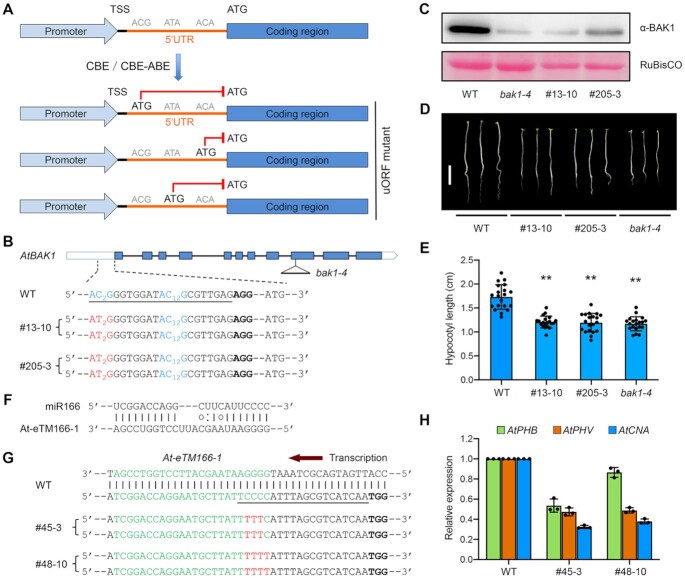
Figure 5.
Repurposing base editors as loss-of-function tools for studying protein-coding or non-coding genes. (A) Schematic diagram illustrating the gene knockdown strategy through creating a uORF in the 5′ UTR of the target gene by base editing. TSS, transcription start site. (B) Two homozygous mutant alleles of Arabidopsis BAK1 containing AID10-nCas9-UGI-created uORF in the 5′ UTR. The target sequence is underlined and the PAM is in bold. The ACG trinucleotides are in blue with subscripted numbers indicating the protospacer positions of target cytosines. (C) Immunoblot analysis indicates significantly decreased AtBAK1 abundances in bak1 mutant alleles relative to wild-type (WT) plants. The T-DNA insertion null mutant bak1-4 was used for reference. Note that the basal immunoblot signals were due to the cross-reactivity of the anti-BAK1 antibody to AtBKK1, a AtBAK1 paralog. RuBisCO, loading control. (D, E) Bak1 5′ UTR mutant seedlings resemble T-DNA insertion null seedlings. The photograph in (D) was taken at five days post germination. Scale bar = 0.5 cm. Quantification of hypocotyl lengths in (E) is presented as means ± s.d. of 20 seedlings. ** P< 0.01 (two-tailed Student's t test). (F) Predicted base pairing between miR166 and the miRNA binding site of At-eTM166-1 in Arabidopsis. (G) Two homozygous mutant alleles of the At-eTM166-1 locus generated by AID10-nCas9-UGI. The miR166 binding site is in green. The target sequence is underlined and the PAM is in bold. Cytosine conversions in mutant lines are in red. (H) qRT-PCR analysis reveals reduced transcript levels of miR166 target genes in At-eTM166-1 mutant lines. Data are presented as means ± s.d. of three biological replicates.