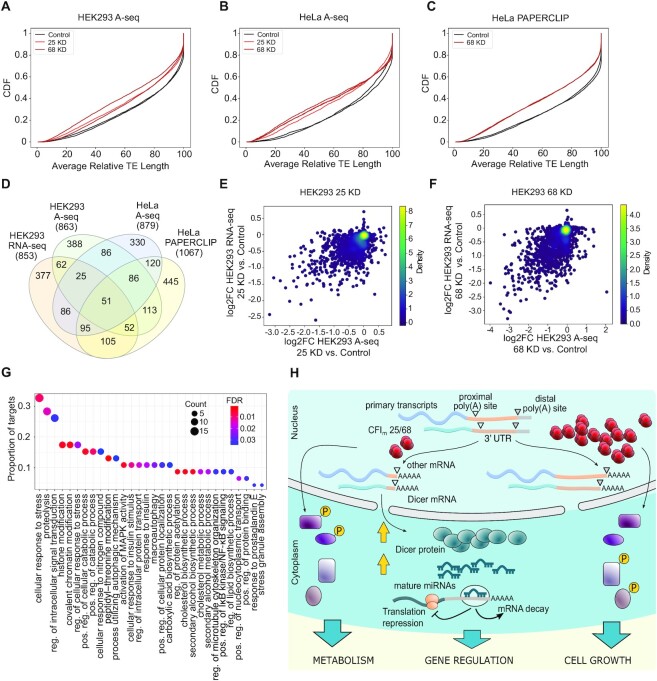
Figure 5.
Overlap of CFIm targets identified with several methodologies from different cell types. (A, B) CDF of average relative TE length inferred from A-seq data in HEK293 cells (A) and HeLa cells (B). The P-value in the two-sample KS-test comparing the HEK293 CFIm25 KD and Control conditions is 3.04e−23, while for CFIm68 KD and Control conditions is 1.70e−43. The P-value for the two-sample KS-test comparing the HeLa CFIm25 KD and Control conditions is 8.75e−22, while for CFIm68 KD and Control conditions is 1.52e−28. (C) CDF of average relative TE length inferred from PAPERCLIP data in HeLa cells; the P-value of the two-sample KS-test comparing HeLa CFIm68 KD and Control conditions is 1.60e−58. (D) Venn diagram (94) showing the overlap of targets obtained from HEK293 RNA-seq, HEK293 A-seq, HeLa A-seq and HeLa PAPERCLIP data sets. The P-value of the overlap of 51 genes among all data sets is 3.66e-101 (SuperExactTest (63)). (E) 2D density plot of log2FC in average relative TE length measured with A-seq or RNA-seq in CFIm25 KD relative to Control cells. The Pearson and Spearman correlation coefficients are, respectively, 0.53 and 0.46 (P-values 6.27e−109 and 3.94e−81). (F) 2D density plot of log2FC in average relative TE length measured with A-seq or RNA-seq in the CFIm68 KD compared to Control cells. Pearson and Spearman correlation coefficients are, respectively, 0.59 and 0.54 (P-values 1.41e−142 and 3.09e−112), (G) Gene Ontology enrichment analysis with the clusterProfiler R package (46) identifies biological processes that are significantly enriched in CFIm targets that are common among 4 cell lines and 2 techniques for quantifying PAS usage (FDR < 0.05). The y-axis shows the proportion of CFIm targets with a specific biological process annotation and the size of the circle is proportional to the absolute number. The color indicates the significance of the enrichment (FDR value). (H) A graphical model of CFIm's impact on several cellular processes.