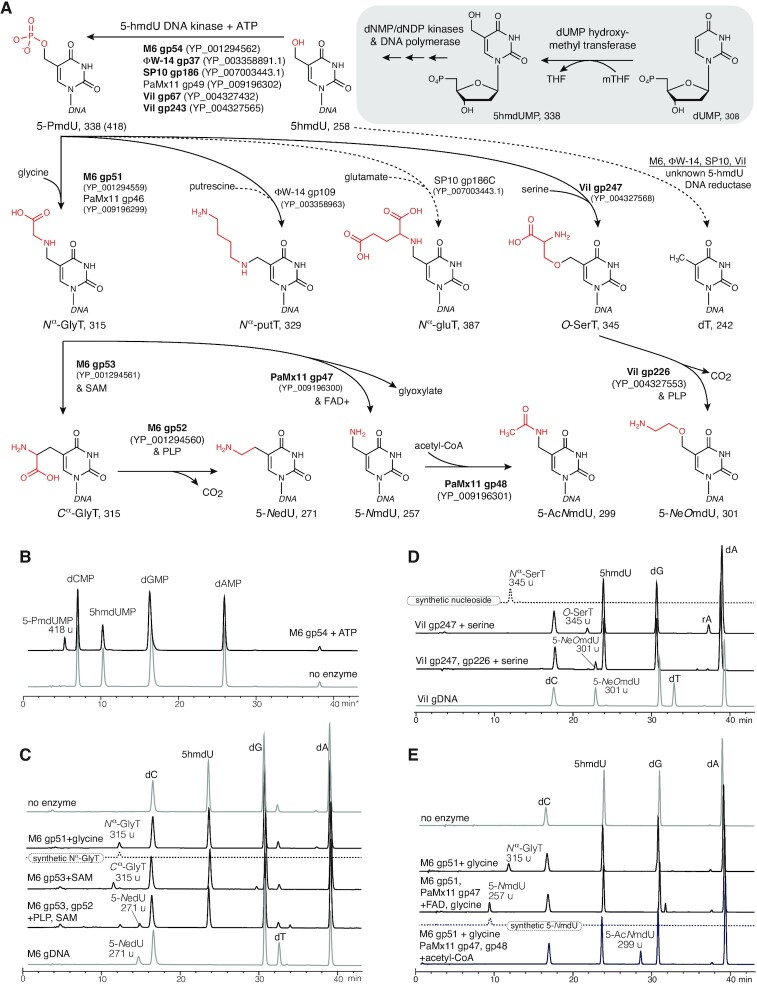
Figure 1.
Thymidine hypermodification pathways, intermediates, cofactors, and products. Pathways of thymidine hypermodification (A). The thymidine hypermodifications discussed in this work utilize 5-hmdU, which is incorporated into DNA through steps occurring before and during DNA replication (as diagrammed in the grayed box) and proceed via a 5-PmdU common intermediate. Solid arrows and bolded enzyme names indicate experimentally verified activities, parentheses contain accession IDs for enzymes, and predicted molecular weights follow the abbreviations for the indicated nucleotide/nucleoside products. (B) HPLC/MS traces of nucleotide mixtures derived from mock treated 5-hmdU substrate DNA (lower trace) and M6 gp54 and ATP. Note, these samples were prepared by enzymatic hydrolysis of DNA in the absence of phosphatase activity. (C) HPLC/MS traces of nucleoside mixtures derived from reactions of 5-hmdU with M6 NedU biosynthetic enzymes and cosubstrates. Traces of no enzyme substrate DNA, synthetic Nα-GlyT and native M6 gDNA included for comparison. (D), HPLC/MS traces of nucleoside mixtures derived from reactions of 5-hmdU with ViI 5-NeOmdU biosynthetic enzymes. Traces from synthetic Nα-SerT (an isomer of O-SerT) and native ViI genomic DNA nucleosides included for comparison to enzymatically produced intermediates and final products, respectively. (E) HPLC/MS traces of nucleoside mixtures derived from reactions with PaMx11 5-AcNmdU biosynthetic enzymes. No enzyme DNA substrate control, and synthetic 5-NmdU standard shown for comparison.