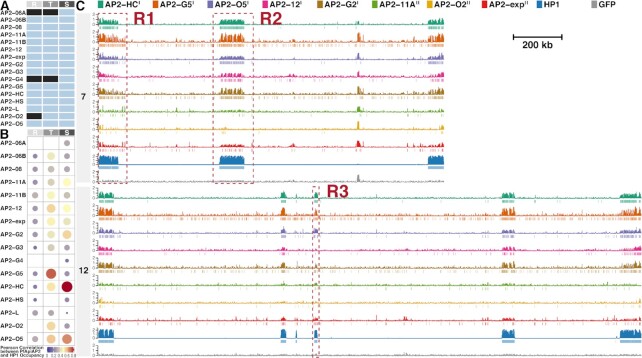
Figure 2.
Eight ApiAP2 transcription factors are associated with heterochromatin. (A) Overview of ChIP-seq assays for profiling ApiAP2 occupancies. Unqualified ChIP-seqs are marked in black. (B) Similarity between ApiAP2 transcription factors and PfHP1 in the genomic occupancy evaluated by Pearson correlation. (C) Log2-transformed ChIP/input fold enrichment and peaks of ApiAP2 transcription factors HFs and PfHP1 in chromosomes 7 and 12 as representatives at the schizont stage except for PfAP2-G5 at the trophozoite stage. In addition, log2-transformed ChIP/input fold enrichment of a GFP negative control is displayed at the bottom for each chromosome. Negative values of log2-transformed ChIP/input fold enrichment are not displayed to present succinct occupancy information. In the legend, PfAP2-HF-Is and PfAP2-HF-IIs are labeled with superscript I and II on the right side, respectively. The genomic regions framed in the red boxes (R1, R2 and R3) are further exemplified in Supplementary Figure S2. R, ring; T, trophozoite; S, schizont.