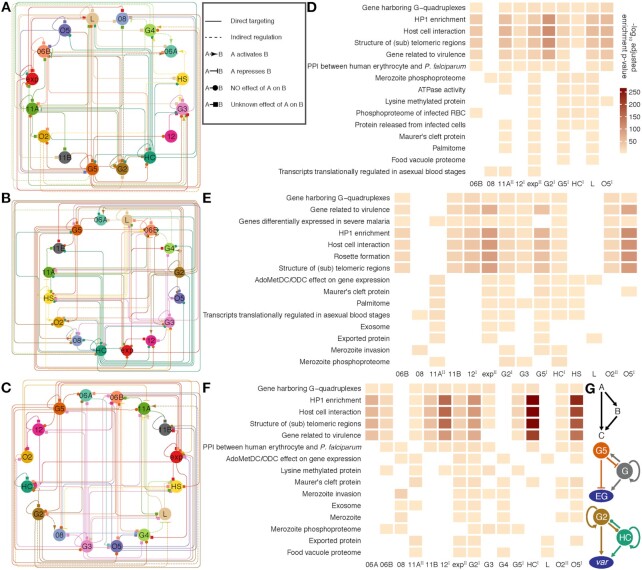
Figure 8.
ApiAP2 regulatory networks in the intraerythrocytic cycle. (A-C) A relationship map of ApiAP2s based on ChIP-sequencing of these transcription factors at the ring, trophozoite, and schizont stages, respectively. ApiAP2 effects on transcription of their targets are indicated if RNA-seq assays of ApiAP2 knockouts are available. Types of relationship are listed in the right box beside (A). (D-F) Representative malaria parasite metabolic pathways (MPMP) targeted by ApiAP2 transcription factors at the ring, trophozoite, and schizont stages, respectively (BH adjusted enrichment P-values < 0.01). The color map is displayed beside (D). PfAP2-HF-Is and PfAP2-HF-IIs are labeled with superscript I and II on the right side, respectively. (G) ApiAP2 transcription factors collaborate via multi-output feedforward loops. Top panel: a simple positive feedforward loop consisting of two transcription factors A and B and a target C. Middle panel: PfAP2-G5 represses pfap2-g transcription, while PfAP2-G activates pfap2-g5 transcription. In addition, PfAP2-G regulates the expression of itself. PfAP2-G5 and PfAP2-G exert opposing effects on expression of early gametocyte (EG) genes. Bottom panel: PfAP2-G2 and PfAP2-HC target each other but do not seem to determine the expression level of the other. Autoregulation exists in both of these two transcription factors. PfAP2-G2 and PfAP2-HC co-activate var transcription.