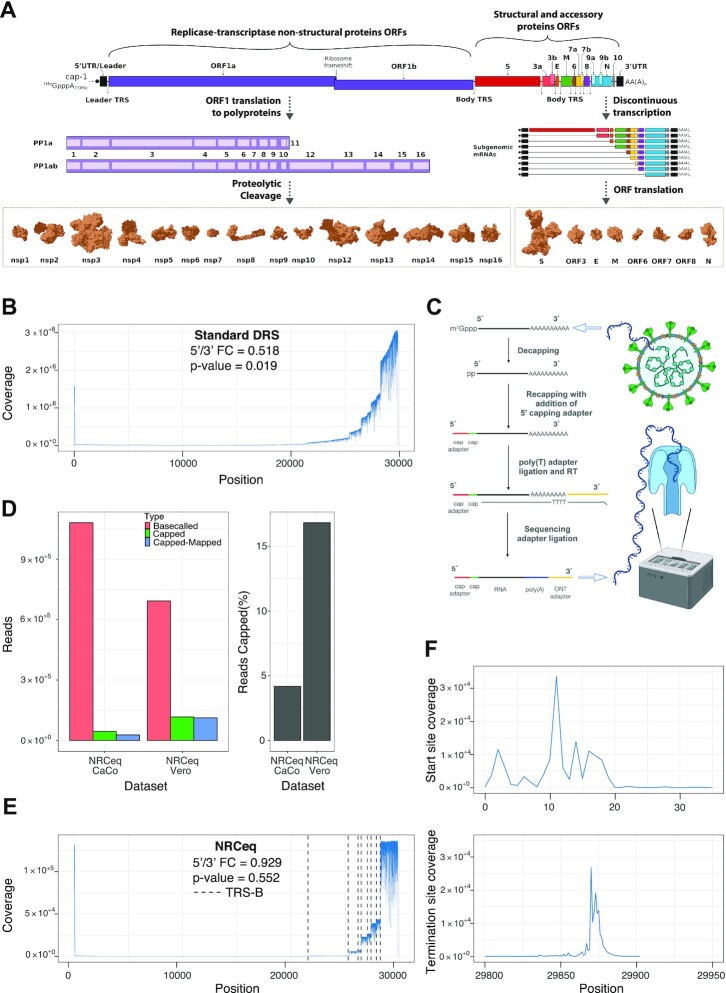
Figure 1.
NRCeq allows sequencing of full-length viral sgRNAs. (A) Schematic representation of the landscape of the SARS-CoV-2 transcriptome, ORFs cleavage sites and protein structures (Adrien Leger (2020): SARS-COV-2 replication cycle. Doi: 10.6084/m9.figshare.12229013.v1). (B) Read coverage across the viral genome calculated from the aggregated standard Nanopore DRS datasets used in this study (see Supplementary Table S1). The figure also reports the coverage fold change between the 5′ (from 15 to 60) and 3′ (region from 29805 to 29850). The reported p-value was calculated with the two-sided Welch's test. (C) Schematic overview of the NRCeq recapping protocol. (D) Number (left) and percentage (right) of basecalled, trimmed and mapped reads for the NRCeq datasets. (E) Read coverage across the viral genome calculated using aggregated NRCeq data from CaCo2 and Vero cells.The figure also reports the coverage fold change between the 5′ (genomic region from 15 to 60) and 3′ (genomic region from 29805 to 29850). The reported p-value was calculated with the two-sided Welch's test. (F) Coverage of the viral genome calculated using only the alignment start sites (top) or alignment termination sites (bottom) for the NRCeq data from CaCo2 and Vero cells, aggregated in a single dataset.