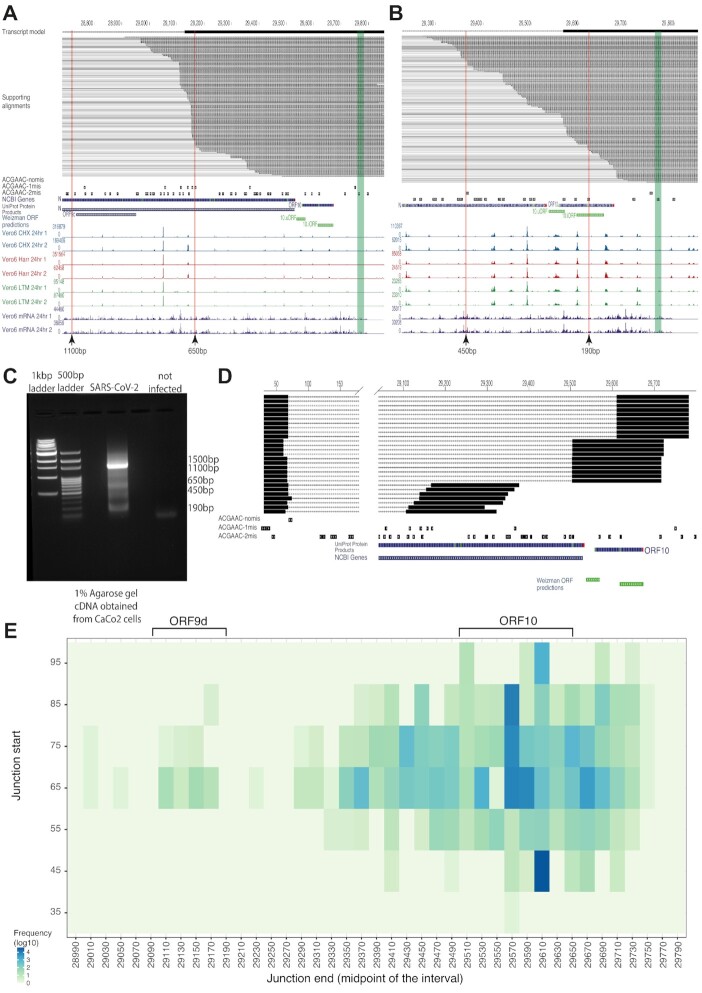
Figure 3.
Independent sgRNAs encode ORF9d and ORF10. (A, B) UCSC Genome Browser track showing the alignments of NRCeq reads assigned to ORF9d (A) and ORF10 (B). The figure also includes tracks showing the location of TRS-B sequences with 0, 1 or 2 mismatches, NCBI Genes, Uniprot Protein Products, ORF predictions and ribosome footprints (46). Arrows indicate the genomic position of the products found in the bands of the gel in (C). (C) Agarose gel electrophoresis after PCR amplification of short sgRNA species. The band at 1500nt, 650nt and 190nt correspond to the expected size for the amplicons of full-length N ORF9d and ORF10 respectively. The bands at 450 and 1100 did not correspond to the size of any assembled transcript models. (D) Representative alignments of Illumina DNA sequencing data (250nt × 2) of short, PCR-amplified sgRNAs. The bands at 1500nt, 650nt, 450nt and 190nt from the gel in (C) were purified and sequenced. (E) Heatmap showing the location and abundance of split alignments connecting the viral 5′UTR with downstream regions. The figure was generated using the Illumina DNA sequencing data as in (D). The y-axis reports the genomic coordinate upstream of the junction, whereas the x-axis reports the genomic coordinate downstream of the junction. The colour scale reports the number of reads that support each junction (log10 transformed) after binning the genome in intervals of 10nt (x-axis) or 20nt (y-axis). Axis labels report the midpoint of the interval.